- Title
-
Nitrate exposure reprograms hepatic amino acid and nutrient sensing pathways prior to exercise: A metabolomic and transcriptomic investigation in zebrafish (Danio rerio)
- Authors
- Keller, R.M., Beaver, L.M., Prater, M.C., Truong, L., Tanguay, R.L., Stevens, J.F., Hord, N.G.
- Source
- Full text @ Front Mol Biosci
|
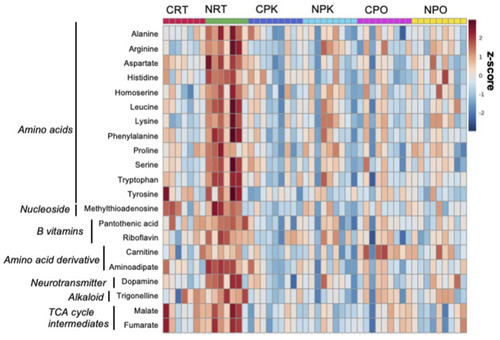
Nitrate treatment altered abundance of metabolites at rest, compared to control liver at rest. The metabolites were chosen based on top FDR-corrected |
|
Metabolites related to NO homeostasis in zebrafish. Relative levels of metabolites metabolized by nitric oxide synthase (NOS), arginase (ARG) and xanthine oxidoreductase (XOR). Citrulline can also be recycled back to arginine. Asl, argininosuccinate lyase; Ass, argininosuccinate synthase. Labeled means without a common letter differ. (One-way ANOVA with Fisher’s post-hoc and Holm FDR-correction, |
|
Nitrate exposure increased metabolites involved in amino acid metabolism, fatty acid metabolism, and the TCA cycle in liver. Labeled means without a common letter differ. (One-way ANOVA with Fisher’s post-hoc and Holm FDR-correction, |
|
Nitrate exposure increased metabolites in the dopamine synthesis pathway. Labeled means without a common letter differ. (One-way ANOVA with Fisher’s post-hoc and Holm FDR-correction, |
|
Expression of genes involved in liver energy metabolism and redox signaling resulting from nitrate treatment. The heat map was generated with Prism and color mapping represents the mean of each treatment group. Negative and positive values indicate downregulation and upregulation compared to control values, respectively. Asterisks indicate significance; *, |