- Title
-
A cAMP Sensor Based on Ligand-Dependent Protein Stabilization
- Authors
- Sidoli, M., Chen, L.C., Lu, A.J., Wandless, T.J., Talbot, W.S.
- Source
- Full text @ ACS Chem. Biol.
|
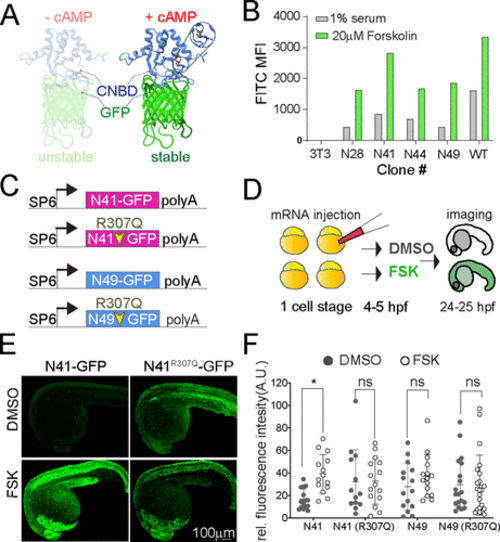
Figure 1. (A) Ribbon diagram of the cAMP sensor composed of CNBD-GFP protein that is unstable without cAMP but stabilized upon cAMP binding: solution structure of a bacterial cyclic nucleotide-activated K+ channel binding domain in cAMP-free form on the left (PBD 2KXL) and bound to cAMP on the right (PBD 2K0G). (B) NIH 3T3 cells stably expressing CNBD-GFP derived from error-prone PCR were treated with 1% serum or 20 μM forskolin for 17 h. (C) Synthetic mRNA encoding the sensor variants N41 and N49 CNBD and the corresponding cAMP-insensitive controls (R307Q). (D) mRNA was injected into zebrafish embryos at the one-cell stage. Embryos were treated at 4–5 hpf with FSK or DMSO for 20 h, then imaged at 24–25 hpf. (E) Representative images showing 24 hpf embryos injected with N41-GFP and N41R307Q-GFP mRNA at one-cell stage and treated with DMSO or 20 μM FSK starting at 4 hpf. (F) Each dot represents mean GFP intensity of one embryo. Error bars indicate SD: *p < 0.5 by one-way ANOVA (with Šídák’s multiple comparisons), n = 13–22 embryos each condition; ns = not significant. AU = arbitrary unit. |
|
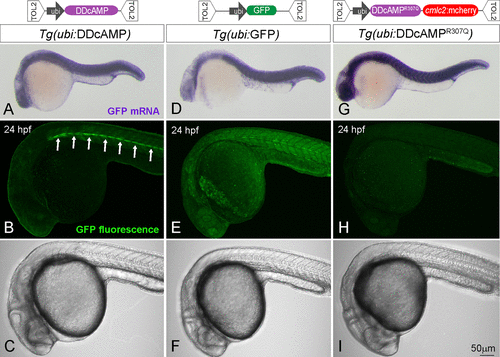
Figure 2. (top) Diagram of the TOL2 plasmids containing N41-GFP, N41R307Q-GFP, and GFP DNA under control of ubi promoter. (bottom) In situ hybridization for GFP mRNA in Tg(ubi:DDcAMP) (A), Tg(ubi:GFP) (D) and Tg(ubi:DDcAMPR307Q) (G) embryos at 24 hpf. Confocal acquisition of GFP signal in Tg(ubi:DDcAMP) (B, C), Tg(ubi:GFP) (E, F), and Tg(ubi:DDcAMPR307Q) (H, I) embryos at 24 hpf. Horizontal myoseptum is indicated by white arrows in panel B. |
|
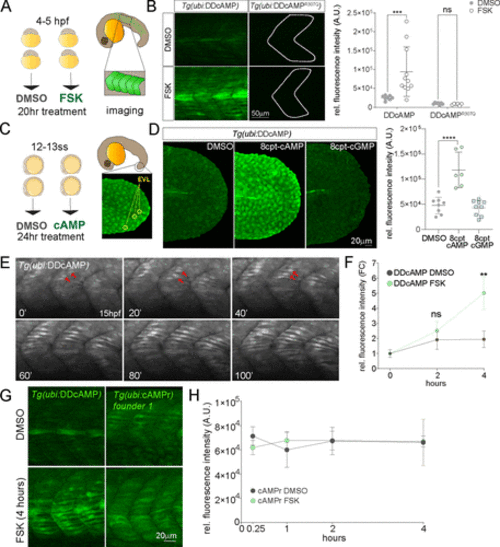
Figure 3. (A) Diagram of DMSO and 20 μM FSK treatment on Tg(ubi:DDcAMP) and Tg(ubi:DDcAMPR307Q) embryos beginning at 4–5 hpf for 20 h. Anterior somites from treated embryos were imaged at 24 hpf, and GFP intensity was measured in somites 7–11. (B) Images of somites from DMSO and FSK-treated Tg(ubi:DDcAMP) and Tg(ubi:DDcAMPR307Q) embryos at 24 hpf. The graph represents the mean fluorescence intensity of five somites per embryo, and each point corresponds to one embryo. Error bars indicate SD: ***p < 0.001 by two-way ANOVA (Šídák’s multiple comparisons), n = 6–11 animals for each condition. ns = not significant. AU = arbitrary unit. (C) Diagram of DMSO and 8-cpt-cAMP treatment of Tg(ubi:DDcAMP) starting at 12–13 somite stage (12–13ss) and imaged after 24 h of incubation. The tip of the tail was imaged for GFP intensity and the EVLs were quantified. (D) Tg(ubi:DDcAMP) embryos from the same clutch were treated with DMSO, 100 μM 8-cpt-cAMP, or 100 μM 8-cpt-cGMP; signal from 20 EVL cells was averaged per animal. Each dot in the graph represents one animal. Error bars indicate SD; ****p < 0.0001 one-way ANOVA (with Bonferroni’s multiple comparisons), n = 6–9 animal per condition. AU = arbitrary unit. (E) Frames from a confocal time lapse image with Airyscan 2 processing of Tg(ubi:DDcAMP) embryo starting at 15 hpf (t = 0 min). One frame every 20 min is shown as representation of the time lapse. GFP-expressing muscle cells are indicated with red arrows. (F) Graph represents the fold change of mean fluorescence intensity measured in five somites per embryo at time 0 and after 2 and 4 h of treatment. Error bars indicate SEM; **p < 0.01 two-way ANOVA (with Šídák’s multiple comparisons), n = 6–10 animals for each condition. ns = not significant, FC = fold change. (G) Confocal images of somites from Tg(ubi:DDcAMP) and Tg(ubi:cAMPr) embryos after 4 h of FSK and DMSO treatment show GFP expression in muscle cells but different response to FSK treatment. (H) Graph represents the mean fluorescence intensity measured in five somites per embryo at the time points indicated. Error bars indicate SEM; there is no significance among the groups; n = 4–5 animals for each condition. EXPRESSION / LABELING:
PHENOTYPE:
|
|
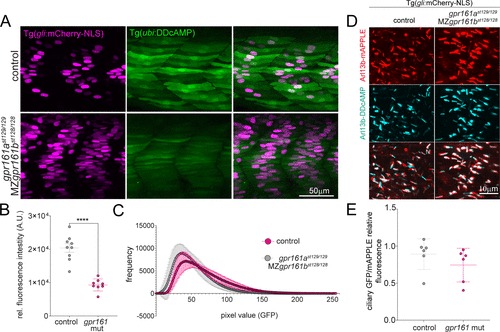
Figure 4. (A) Confocal images of cAMP (green) and Shh reporter (magenta) signals in control fish and double mutants forgpr161a (st129 allele) and MZgpr161b (st128 allele). MZ = maternal zygotic. Control fish include gpr161ast129/+, gpr161b128/+ and gpr161ast129/129, gpr161b128/+. (B) Graphs show GFP fluorescence intensity measured in the somite area corresponding to the muscle pioneers in Tg(gli:mCherry-NLS); Tg(ubi:DDcAMP) (control) and in Tg(gli:mCherry-NLS); Tg(ubi:DDcAMP); gpr161ast129/129; MZgpr161b128/128 (gpr161 mut) embryos. Error bars indicate SD: ****p < 0,0001 by t-test. (C) Mean of the pixel values in each image acquired in panel A plotted as a function of their frequency. gpr161 double mutant fish display darker values compared to the controls. n = 4–5 animals per genotype. (D) Magnification of somites from 24 hpf Tg(gli:mCherry-NLS) control and Tg(gli:mCherry-NLS); gpr161ast129/129; MZgpr161b128/128 embryos injected at one-cell stage with Arl13b-DDcAMP (cyan) and Arl13b-mApple (red cilia). (E) Ratio of ciliary GFP/ciliary mAPPLE in over 20 cilia per animal in Tg(gli:mCherry-NLS) (control) and Tg(gli:mCherry-NLS); gpr161ast129/129; MZgpr161b128/128 (gpr161 mut) embryos. |