- Title
-
Polygoni multiflori radix extracts inhibit SARS-CoV-2 pseudovirus entry in HEK293T cells and zebrafish larvae
- Authors
- Wang, X., Lin, S., Tang, R.W., Lee, H.C., Chan, H.H., Choi, S.S.A., Leung, K.W., Webb, S.E., Miller, A.L., Tsim, K.W.
- Source
- Full text @ Phytomedicine
|
HPLC chromatograms of the PMR extracts. The characteristic peaks of mixed standard markers, and those found in PMRwater and PMREtOH, which were revealed at an absorbance of 230 nm. Insert shows data at higher mAU. 1: EGCG; 2: THSG; 3: emodin; 4: physcion. |
|
Inhibition of S-protein-hACE2 binding. S-protein-hACE2 binding was analysed by ELISA. (A) Line graph showing the response of a standard inhibitor (calibrated to NIBSC code 20/136), which was used as a positive control. (B) S-protein-hACE2 binding was inhibited by PMRwater and PMREtOH in a dose-dependent manner. The inhibition percentage was determined from the binding signal normalized to the interaction between spike RBD and hACE2 without extract. The data represent mean ± SD ( |
|
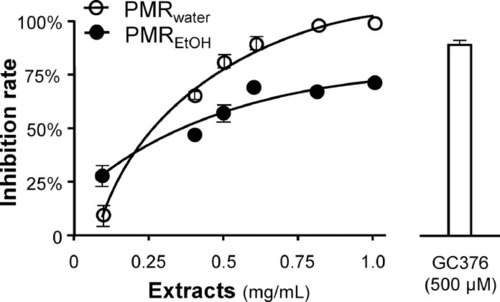
Inhibition of 3CL protease activity. PMRwater and PMREtOH inhibited the activity of 3CL protease of SARS-CoV-2 in a dose-dependent manner. A control inhibitor, GC-376 (500 μM), was used as a positive control. The inhibition percentage was determined according to the optical density normalized to the 3CL protease activity without extract. The data represent mean ± SD ( |
|
Inhibition of pseudovirus entry into HEK293T cells overexpressing hACE2. (A) The amount of intracellular luciferase activity (indicating the rate of virus entry) was measured at different dilutions of the pseudovirus. (B) Pseudovirus entry was inhibited by PMRwater or PMREtOH in a dose-dependent manner. The inhibition percentage was determined according 20 to the luciferase activity normalized to the luciferase activity without extract. A SARS-CoV-2 neutralizing antibody was used as a positive control. The data represent mean ± SD, ( |
|
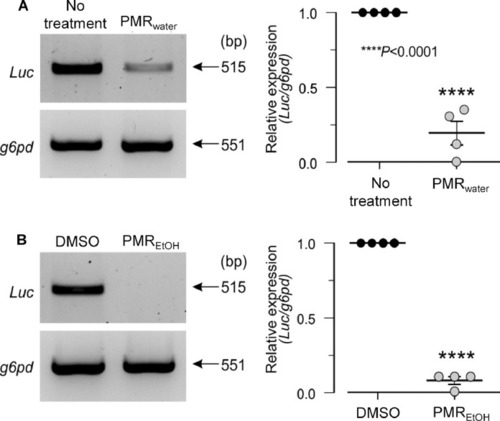
PMRwater and PMREtOH inhibit the entry of SARS-CoV-2 PEG-pseudovirus in zebrafish. Larvae at 3 dpf were pre-treated with (A) PMRwater or (B) PMREtOH, then co-treated with the respective extract and SARS-CoV-2 PEG-pseudovirus for a further 72 h. PCR amplification of whole larval cDNA showed the relative level of Luc expression in the treatment and control groups (left). The relative level of Luc expression was quantified by normalization to the expression level of g6pd (right). The data show that treatment with PMRwater or PMREtOH inhibited the expression of Luc (and thus the entry of PEG-pseudovirus) in larvae. The data represent the mean ± SD ( |
|
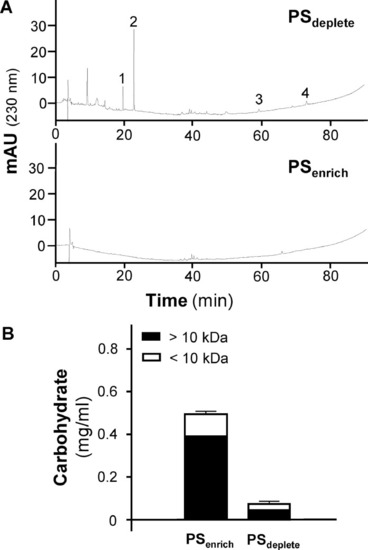
Fractionation of PSenrich and PSdeplete. (A) HPLC analyses of both fractions, at 1 mg/ml, were revealed at 230 ± 4 nm. EGCG (1), THSG (2), emodin (3) and physcion (4) are marked. (B) PSdeplete and PSenrich (0.5 ml each at 1 mg/ml dried weight) were transferred to a 10 kDa cut-off concentrator (Sartorius, German) and centrifuged at 14,000 g for 15 min. The carbohydrate content was measured with phenol-sulfuric acid. The values are expressed as mean ± SD, |
|
Inhibition of pseudovirus entry by PSenrich, PSdeplete and EGCG. Pseudovirus entry in hACE2-overexpressing HEK293T cells were inhibited by (A) PSenrich and PSdeplete, and (B) EGCG, all in a dose-dependent manner. The data represent mean ± SD, ( |
Reprinted from Phytomedicine : international journal of phytotherapy and phytopharmacology, 102, Wang, X., Lin, S., Tang, R.W., Lee, H.C., Chan, H.H., Choi, S.S.A., Leung, K.W., Webb, S.E., Miller, A.L., Tsim, K.W., Polygoni multiflori radix extracts inhibit SARS-CoV-2 pseudovirus entry in HEK293T cells and zebrafish larvae, 154154, Copyright (2022) with permission from Elsevier. Full text @ Phytomedicine