- Title
-
Bioluminescent Zebrafish Transplantation Model for Drug Discovery
- Authors
- Hason, M., Jovicic, J., Vonkova, I., Bojic, M., Simon-Vermot, T., White, R.M., Bartunek, P.
- Source
- Full text @ Front Pharmacol
|
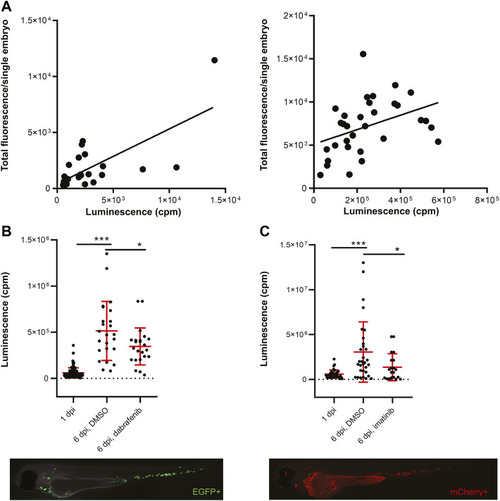
Transplanted cancer cells survive |
|
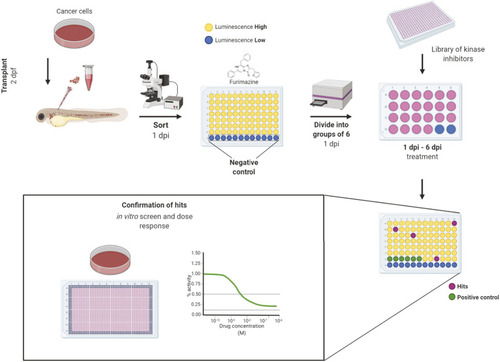
Workflow of |
|
Kinase inhibitors active in transplanted ZMEL1 melanoma cells. |
|
Kinase inhibitor active in transplanted leukemia K562 cells. |
|
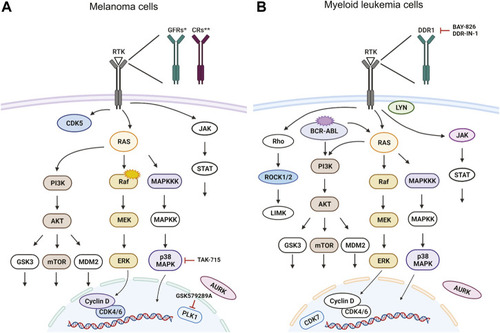
Targeted signaling pathways as predicted from |