- Title
-
Fibroblast growth factor receptor 4 promotes glioblastoma progression: a central role of integrin-mediated cell invasiveness
- Authors
- Gabler, L., Jaunecker, C.N., Katz, S., van Schoonhoven, S., Englinger, B., Pirker, C., Mohr, T., Vician, P., Stojanovic, M., Woitzuck, V., Laemmerer, A., Kirchhofer, D., Mayr, L., LaFranca, M., Erhart, F., Grissenberger, S., Wenninger-Weinzierl, A., Sturtzel, C., Kiesel, B., Lang, A., Marian, B., Grasl-Kraupp, B., Distel, M., Schüler, J., Gojo, J., Grusch, M., Spiegl-Kreinecker, S., Donoghue, D.J., Lötsch, D., Berger, W.
- Source
- Full text @ Acta Neuropathol Commun
|
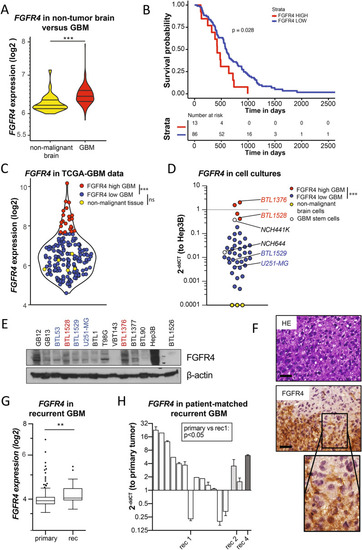
FGFR4 is overexpressed in a highly aggressive GBM subgroup, associated with tumor recurrence. |
|
|
|
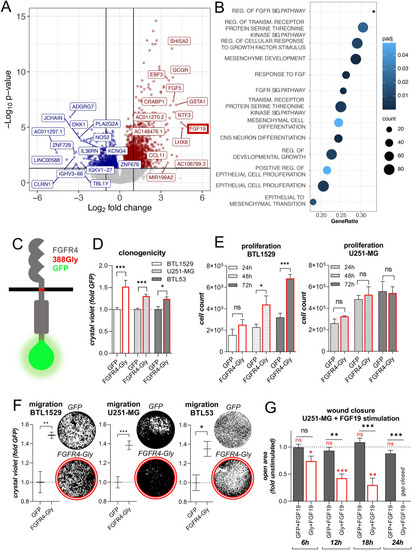
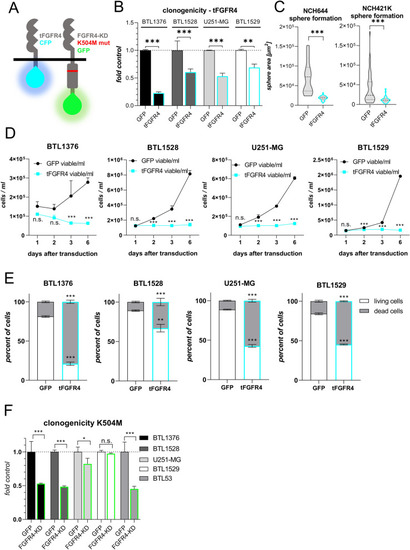
Inactivation of FGFR4 reduces proliferative capacity and promotes cell death. |
|
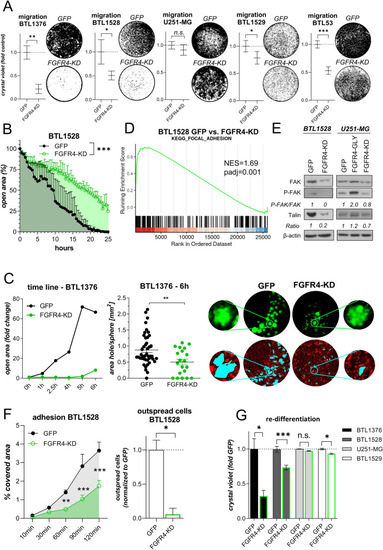
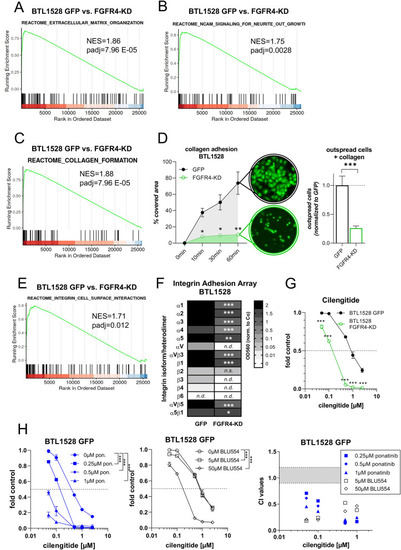
FGFR4 inactivation attenuates GBM cell migration, endothelial invasion, and adhesion. |
|
FGFR4 blockade results in loss of GBM cell adhesion and sensitizes towards the |
|
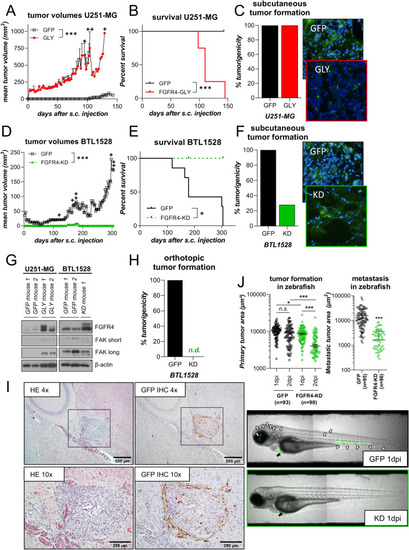
FGFR4 impacts on tumorigenicity and tumor growth. PHENOTYPE:
|