- Title
-
The Role of Microtubule Associated Serine/Threonine Kinase 3 Variants in Neurodevelopmental Diseases: Genotype-Phenotype Association
- Authors
- Shu, L., Xiao, N., Qin, J., Tian, Q., Zhang, Y., Li, H., Liu, J., Li, Q., Gu, W., Wang, P., Wang, H., Mao, X.
- Source
- Full text @ Front. Mol. Neurosci.
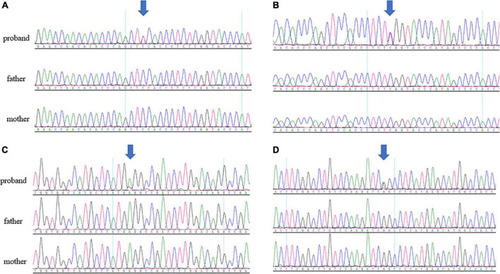
|
|
|
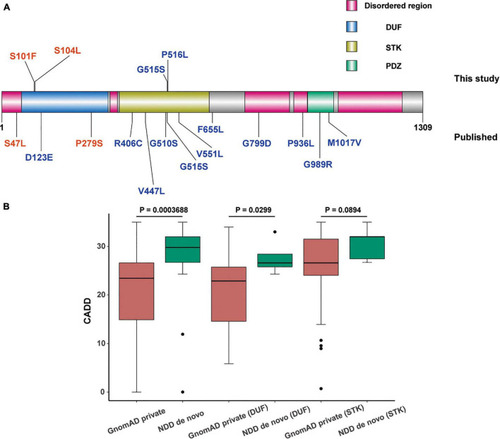
Distribution and comparison of pathogenicity of missense mutations in domain of unknown function (DUF) and serine-threonine kinas (STK) domain of |
|
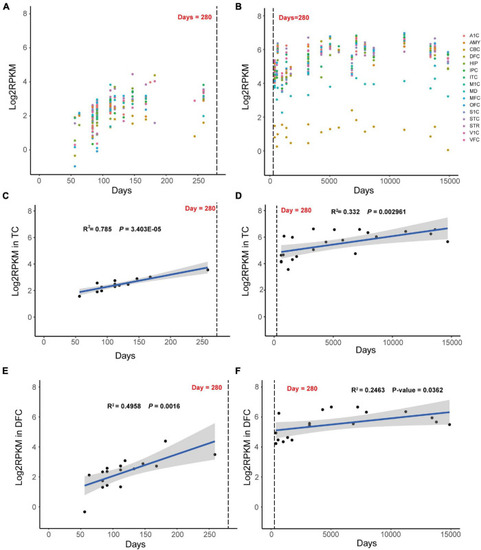
Dynamic expression mode of |
|
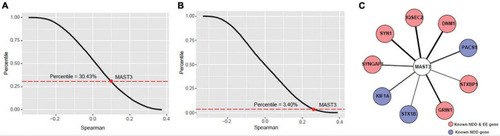
Co-expression analyses of microtubule associated serine/threonine kinase 3 (MAST3) with EE and NDD genes. |
|
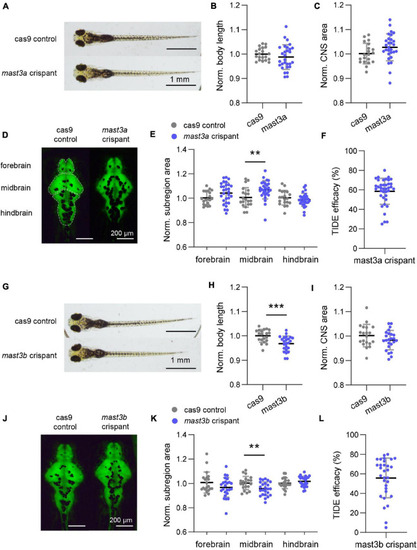
Disruption of zebrafish mast3a and mast3b led to abnormal central nervous system (CNS) morphology. (A) Representative bright-field imaging of larval zebrafish at six dpf (dorsal view). Top, cas9 injected control; bottom, mast3a F0 CRISPR (crispant). (B) Measurements of body length in cas9 injected control (n = 20 fish) vs. mast3a crispant (n = 29 fish). Data were normalized to the average body length of cas9 control group. (C) Normalized CNS area in cas9 injected controls vs. mast3a crispants. Data were corrected by the normalized body length of individual larvae. (D) Representative imaging of HuC: eGFP expressed larval zebrafish shows CNS fluorescence pattern at six dpf (dorsal view). Left, cas9 injected control; right, mast3a crispant. White dash line highlights the CNS subregions, forebrain, midbrain, and hindbrain, for measurement. (E) Normalized CNS subregion area in cas9 injected control vs. mast3a crispant. Data were normalized to the average of each subregion area of cas9 control group, and then corrected by the normalized body length of individual larvae (gray dot, cas9 control; blue dot, mast3a crispant). (F) CRISPR efficacy calculated via TIDE method of individual mast3a crispant used in phenotypic study (n = 38 fish). (G–L) Data from mast3b crispant study (for imaging study, n = 20 fish for cas9 control, and n = 24 fish for mast3b crispant; n = 31 crispant for TIDE efficacy verification). Scale bars as indicated in the figure. Error bars indicate SD. Statistical significance is indicated as **p < 0.01, and ***p < 0.001. |