- Title
-
Loss of Neuropilin2a/b or Sema3fa alters olfactory sensory axon dynamics and protoglomerular targeting
- Authors
- Cheng, R.P., Dang, P., Taku, A.A., Moon, Y.J., Pham, V., Sun, X., Zhao, E., Raper, J.A.
- Source
- Full text @ Neural Dev.
|
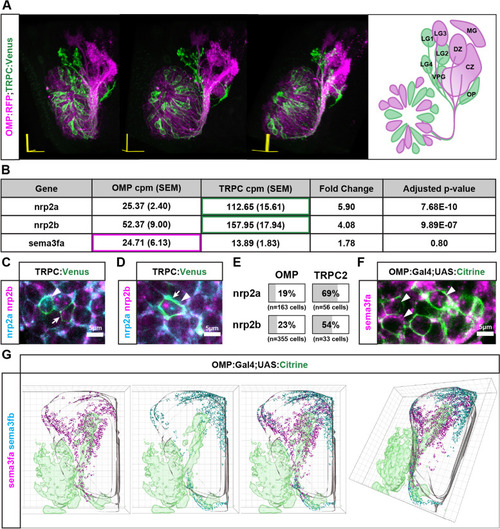
Expression patterns of nrp2a, nrp2b, and sema3fa in the developing zebrafish olfactory system. A. 3D projection and schematic representation (frontal view) of an optical z-stack of the 72 hpf olfactory system. One side of the olfactory system is shown with dorsal up and lateral on the left. OSNs are labeled by OMP:RFP (Magenta) and TRPC2:Venus (Green). OMP-expressing axons project to the central zone (CZ), dorsal zone (DZ), lateral glomerulus 3 (LG3), and medial glomerulus (MG). TRPC2-expressing axons project to the olfactory plexus (OP), lateral glomerulus 1,2, and 4 (LG1, LG2, LG4), and the ventral posterior glomerulus (VPG). B. RNAseq expression data from four replicate experiments for sema3fa, nrp2a, and nrp2b in OMP and TRPC2 expressing OSNs. C. Nrp2a mRNA (cyan, arrowhead) and nrp2b mRNA (magenta, arrow) is detected independently in the cell bodies of TRPC2:Venus labeled cells (green) in the OE at 48 hpf. D. Nrp2a mRNA (cyan, arrowhead) and nrp2b mRNA (magenta, arrow) is detected in the same cell body of TRPC2:Venus labeled expressing cells (green) in the OE at 48 hpf. E. Percentage of OMP and TRPC2 expressing cells with nrp2a or nrp2b expression. F. Sema3fa mRNA (magenta) is detected in the cell bodies of OMP:Gal4;UAS:Citrine expressing cells (green) in the OE at 48 hpf. G. Sema3fa and sema3fb expression in the olfactory system of 36 hpf zebrafish. 3D model of sema3fa mRNA (magenta) and sema3fb mRNA (cyan) distribution within the OB (grey outline). Sema3fa is concentrated in the anterior and dorsal-medial regions of the OB and sema3fb is concentrated in the dorsal-medial regions of the olfactory bulb. Data aggregated from 6 OB hemispheres for sema3fa and 6 OB hemispheres for sema3fb |
|
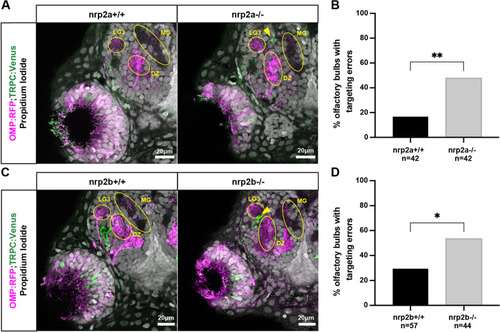
Nrp2a and nrp2b are required for normal protoglomerular targeting of TRPC2-class OSNs. A. Representative confocal sections of wild type and nrp2a mutant siblings. Yellow arrows indicate misprojecting TRPC2-class axons. B. The percentage of olfactory bulbs with targeting errors is higher in nrp2a mutants as compared to wild type siblings. C. Representative confocal sections of wild type and nrp2b mutant siblings. Yellow arrows indicate misprojecting axons. D The percentage of olfactory bulbs with targeting errors is higher in nrp2b mutants as compared to wild type siblings |
|
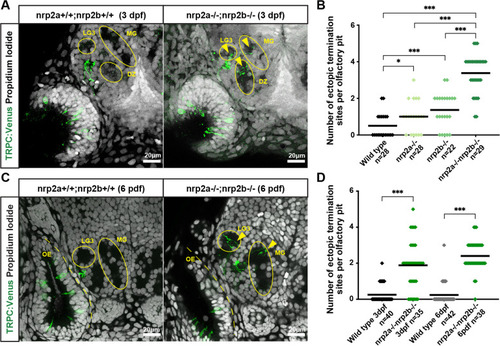
Nrp2a and nrp2b likely work in parallel and targeting errors persist for days. A. Representative confocal sections of 3 dpf wild type and nrp2a;nrp2b double mutant siblings showing multiple misprojections by TRPC2-class OSNs in the MG, DZ, and LG3 protoglomeruli. Yellow arrows indicate misprojecting axons. B. TRPC2-class OSNs in nrp2a;nrp2b double mutants project axons to more ectopic termination sites than single mutant or wild type siblings. C. Representative confocal sections of 6 dpf wild type and nrp2a;nrp2b double mutant siblings showing multiple misprojections by TRPC2-class OSNs in the MG and LG3 protoglomeruli. Yellow arrows indicate misprojecting axons. D Targeting errors of TRPC2-class OSNs persist into later stages of development. The number of ectopic termination sites per olfactory pit is significantly higher in nrp2a;nrp2b double mutant as compared to wild type siblings at both 3 dpf and at 6 dpf |
|
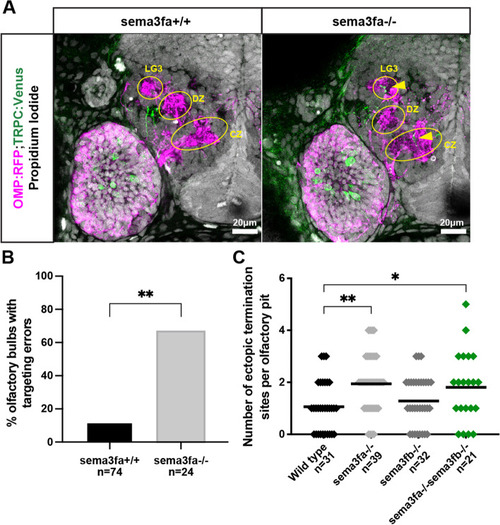
Sema3fa is required for normal TRPC2-class OSN protoglomerular targeting. A. Representative confocal sections of wild type and sema3fa mutant siblings. Yellow arrows indicate misprojecting axons. B. The percentage of olfactory bulbs with targeting errors is higher in sema3fa mutants as compared to wild type siblings. C. The misprojection phenotype of Sema3fa mutants is not significantly different from sema3fa;sema3fb double mutants |
|
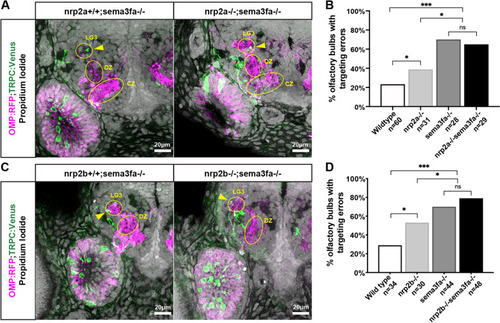
Both nrp2a and nrp2b act in the same pathway with sema3fa. A. Representative confocal sections of sema3fa mutant and nrp2a;sema3fa double mutant siblings. Yellow arrows indicate misprojecting axons. B. The misprojection phenotype in nrp2a;sema3fa double mutants is not significantly different from sema3fa mutant siblings. C. Representative confocal sections of sema3fa mutant and nrp2b;sema3fa double mutant siblings. Yellow arrows indicate misprojecting axons. D. The misprojection phenotype in nrp2b;sema3fa double mutants is not significantly different from sema3fa mutant siblings |
|
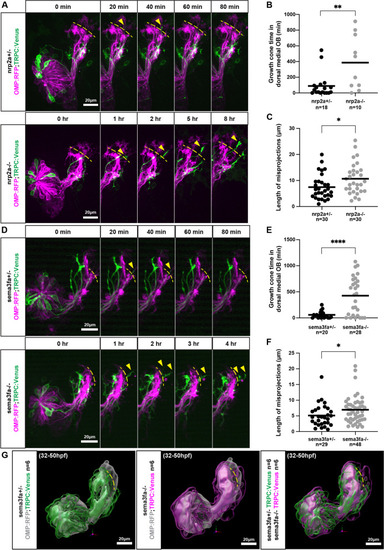
Misprojecting growth cones fail to retract in nrp2a and in sema3fa mutants. A. Live imaging sequences of nrp2a heterozygote and mutant siblings, showing misprojecting axons occupying the dorsal-medial OB. The yellow dotted lines indicate the dorsal boundary of the developing DZ and CZ protoglomeruli and denote the edge of the dorsal-medial OB region. Yellow arrows indicate misprojecting axons. B. The cumulative time that the dorsal-medial OB is occupied by TRPC2-class OSNs is greater in nrp2a mutants as compared to nrp2a heterozygous siblings. C. The maximum distance TRPC2-class axons project into the dorsal-medial OB is greater in nrp2a mutants as compared to heterozygotes. D. Live imaging sequences of sema3fa heterozygote and mutant siblings, showing misprojecting axons occupying the dorsal-medial OB. E. The cumulative time that the dorsal-medial OB is occupied by TRPC2-class OSNs is greater in sema3fa mutants as compared to heterozygous siblings. F. The maximum distance TRPC2-class axons project into the dorsal-medial OB is greater in sema3fa mutants as compared to heterozygotes. G. Model of average TRPC2-class axon locations in sema3fa heterozygotes and mutants during live imaging sequence. TRPC2-class axons are shown in green and magenta, and OMP-class axons in grey. The three TRPC2 surfaces encompass axon location probabilities, from most transparent to most opaque, of 5.6, 18.7, and 31.8%. Yellow dotted line represents the edge of the dorsal-medial OB region |