- Title
-
Glutaredoxin 2 promotes SP-1-dependent CSPG4 transcription and migration of wound healing NG2 glia and glioma cells: Enzymatic Taoism
- Authors
- Wilms, C., Lepka, K., Häberlein, F., Edwards, S., Felsberg, J., Pudelko, L., Lindenberg, T.T., Poschmann, G., Qin, N., Volbracht, K., Prozorovski, T., Meuth, S.G., Kahlert, U.D., Remke, M., Aktas, O., Reifenberger, G., Bräutigam, L., Odermatt, B., Berndt, C.
- Source
- Full text @ Redox Biol.
|
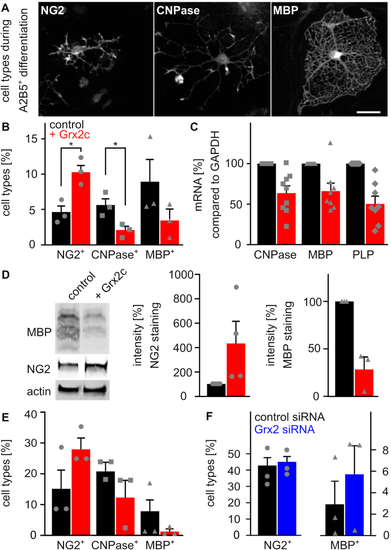
Glutaredoxin 2c inhibits differentiation of mouse oligodendrocyte progenitor cells into mature oligodendrocytes. A) Primary mouse A2B5+ cells differentiate into mature MBP+ oligodendrocytes via NG2−glia and CNPase+ cells. Immunocytochemical analyses were performed after 5 days of differentiation, scale bar: 10 μm. B) Differentiation of A2B5+ cells with or without treatment with 2 μM recombinant Grx2c, quantification after immunohistochemistry from 3 independent biological samples. C) qRT-PCR of indicated genes after 5 days of differentiation of A2B5+ cells using GAPDH for normalization. D) Representative western blot analyses and quantification of 3 independent experiments after 5 days of differentiation of A2B5+ cells. E) Differentiation of NG2+ cells with or without treatment with 2 μM recombinant Grx2c, quantification after immunohistochemistry from 3 independent biological samples. F) qRT-PCR of indicated genes after 5 days of differentiation of NG2+ cells treated either with control siRNA or Grx2siRNA. Data were normalized to transcripts of GAPDH. *: p < 0.05 (Student's t-test). |
|
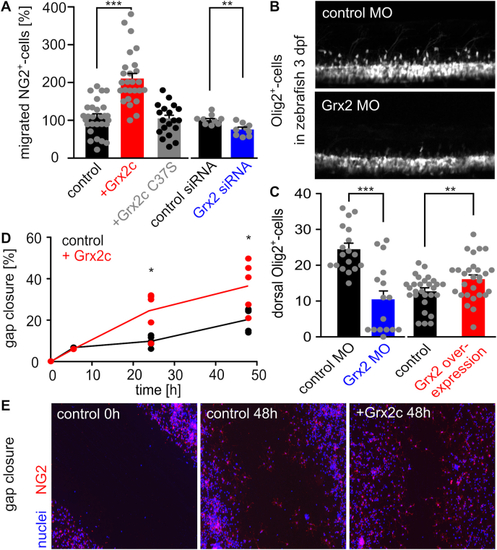
Glutaredoxin 2c increases migratory capacity of mouse NG2+ glia. A) NG2+ cells were seeded in a Boyden chamber and incubated with 2 μM Grx2cwt or Grx2cC37S or transfected either with control siRNA or Grx2siRNA. Transmigrated cells were counted. B) Two-photon-microscopy of dorsally migrating oligodendrocyte progenitor cells in Tg(olig2-GFP) zebrafish at 3 dpf with and without morpholino-induced Grx2 knock-down. C) Migrated cells shown in b) were counted in zebrafish 3 dpf comparing Grx2 knock-down or Grx2 overexpression with respective controls. D) Quantified gap closure after scratch assay at indicated time points by NG2+ cells treated with 2 μM recombinant Grx2c or not. E) Immunocytochemistry of control and Grx2c-treated NG2+ cells after scratch assay at indicated time points. *: p < 0.05; **: p < 0.01; ***: p < 0.001 (Student's t-test). PHENOTYPE:
|
|
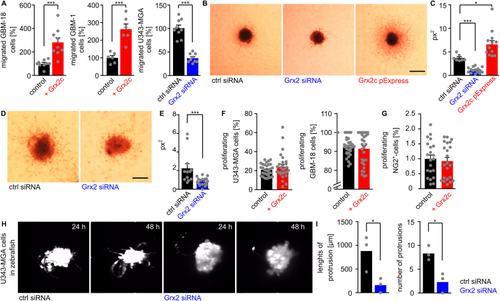
Glutaredoxin 2c promotes invasion of human glioblastoma cells. A) Quantification of transmigrated glioblastoma cells using a Boyden chamber assay, different glioblastoma cell lines, as well as recombinant protein and siRNA to modulate Grx2 levels. B-E) U343-MGA (B) or GBM-18 (D) spheres with modulated Grx2 levels (down-regulation by siRNA, up-regulation by overexpression via a pExpress plasmid) were embedded in a 3D collagen matrix and invasion of cells after 72 h is shown, scale bars: 200 μm. C/E) Quantification of the areas of migrated cells (px2) based on B/D. F,G) U343-MGA, GBM-18 cells (F), and NG2+-cells (G) were treated with or without 2 μM recombinant Grx2c and BrdU+-cells were quantified. H) Fluorescent labelled U343-MGA cells transfected with either control siRNA or Grx2 siRNA were injected into zebrafish at the 1000 cell stage. Pictures show stills from supplemental movies 1 obtained by light sheet microscopy between 24 and 48 hpf. I) Quantification of invasion in 3 zebrafish embryos based on supplemental movies 2. *: p < 0.05; ***: p < 0.001 (Student's t-test). |
|
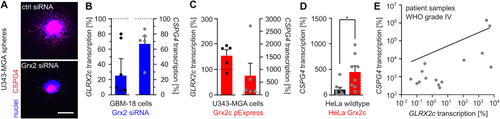
Glutaredoxin 2c regulates CSPG4 transcription. A) Nuclei and CSPG4+ cells were stained in U343-MGA spheres with or without decreased Grx2 levels, scale bar: 200 μm. B) - E) Determination of GLRX2c and CSPG4 transcripts in GBM-18 (B), U343-MGA cells (C), or HeLa cells (D) with and without modulated Grx2 levels and in mRNA isolated from patient samples (E, R2 = 0.358). Data in B) - E) were normalized to GAPDH transcripts. |
|
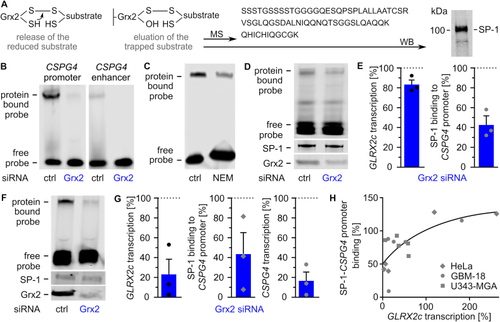
Glutaredoxin 2 acts on CSPG4 transcription and migration via SP-1. A) Scheme of the trapping experiment revealing presence of SP-1 in the eluted substrate after western blot (WB) and mass spectrometry (MS). B) Electrophoretic mobility shift assays (EMSAs) using cell lysates of HeLa cells transfected with either control siRNA or Grx2 siRNA and probes consisting of the promoter or an enhancer region of the CSPG4 gene. C) EMSAs comparing SP-1 probe binding in HeLa lysates treated with or without 100 mM N-ethyl-maleimide. D) EMSAs of GBM-18 cell lysates transfected with either control siRNA or Grx2 siRNA. Shown is the binding of a CSPG4 promoter probe containing SP-1 binding sites during the shift assay and staining of SP-1 as well as Grx2 after western transfer of the same lysate. E) Quantification of D) and qRTPCR was performed to quantify GLRX2c transcription in the cells. F) EMSAs using extracts of HeLa cells transfected with either control siRNA or Grx2siRNA and the probe described in D). The levels of SP-1 and Grx2 were determined after western transfer using the same lysates. G) Quantification of GLRX2c and CSPG4 transcripts, as well as binding between SP-1 and probe in HeLa cells with decreased Grx2 levels compared to control cells. H) SP1-binding to the probe was plotted against GLRX2c transcripts based on all shift assays performed with HeLa, GBM-18, and U343-MGA cell lysates, R2 = 0.543. |
|
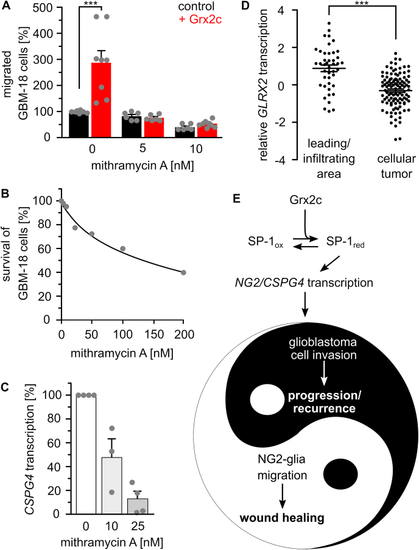
SP-1 is a main target of glutaredoxin 2′s regulation of migration. A) Treatment with 5 and 10 nM of the SP-1 inhibitor mithramycin A abolished the positive effect of Grx2c incubation on GBM-18 cell migration, ***: p < 0.001 (Student's t-test). B) Survival of GBM-18 cells treated with indicated concentrations of mithramycin A was tested using the Cell Titer Blue assay. C) CSPG4 transcripts in GBM-18 cells treated with 10 or 25 nM mithramycin A measured by qRTPCR and normalized to GAPDH transcripts. D) GLRX2 transcripts in the invading areas compared to all other areas of a tumor according to the IVY GAP project. ***: p < 0.001 (ANOVA). E) The Yin and Yang of the (patho-) physiological consequences of Grx2c regulated CSPG4 transcription. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.) |