- Title
-
A highly conserved zebrafish IMPDH retinal isoform produces the majority of guanine and forms dynamic protein filaments in photoreceptor cells
- Authors
- Cleghorn, W.M., Burrell, A.L., Giarmarco, M.M., Brock, D.C., Wang, Y., Chambers, Z.S., Du, J., Kollman, J.M., Brockerhoff, S.E.
- Source
- Full text @ J. Biol. Chem.
|
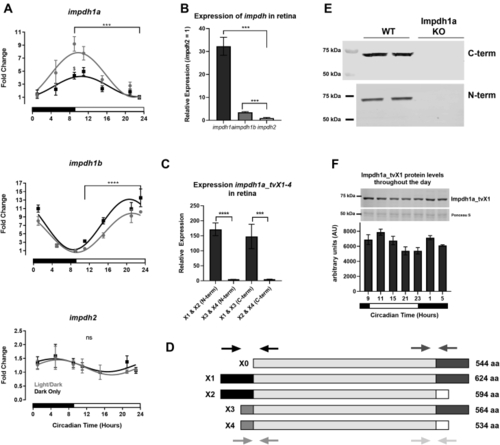
Impdh1a_tv1a is the predominant variant in the retina.A, relative expression levels of all transcript variants for impdh1a, 1b, and 2 throughout the day. For each graph, minimum expression = 1: impdh1a timepoints measured relative to 23:00, impdh1b relative to 9:00, and impdh2 relative to 15:00. The black and white bars indicate lights on (9:00) and lights out (23:00) for animals under normal light/dark cycle, $ denotes p ≤ 0.05 normal light/dark compared with dark only at 9:00. B, comparison of relative maximum expression levels of impdh1a (9:00), 1b (23:00), and 2 (5:00) transcripts. C, comparison of relative expression levels for all impdh1a transcripts detected in the retina at 9:00. N = 6 animals, the samples were in triplicate. D, schematic of impdh1a transcripts X1 to X4 and 544 (X0, NM_001002177.1); primer location for (C) indicated by arrows for both C-terminal and N-terminal reactions. See Table 1 for primer/transcript information. E, Western blot of Impdh1a protein in WT and impdh1a23234 zebrafish retinas using antibodies that recognize either X1 and X3 (C-terminal) or X1 and X2 (N-terminal) N = 2. F, quantification of Impdh1a_tvX1 protein levels throughout the day using the C-terminal antibody, and inset shows representative Western blot with Ponceau S used as a loading control. Trend p =0.04, N = 2 animals per time point. N = 3 animals per time/data point for all other panels except where noted. impdh, inosine monophosphate dehydrogenase; tv, transcript variant. ns p > 0.05, ∗ p ≤ 0.05 , ∗∗ p ≤ 0.01, ∗∗∗ p ≤ 0.001, ∗∗∗∗ p ≤ 0.0001. |
|
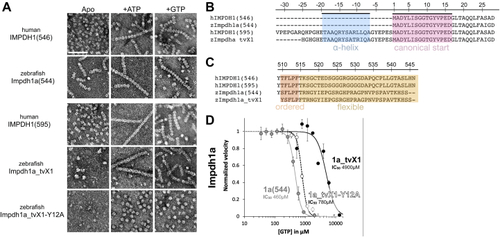
Structural and functional conservation between human and zebrafish IMPDH1.A, negative stain EM of purified human retinal variants IMPDH1(546) & IMPDH1(595) and zebrafish Impdh1a (544) & Impdh1a_tvX1. The scale bar represents 100 nm. B and C, sequence alignment of human retinal variants IMPDH1(546) and IMPDH1(595), zebrafish Impdh1a (544) {"type":"entrez-nucleotide","attrs":{"text":"NM_001002177.1","term_id":"50345007","term_text":"NM_001002177.1"}}NM_001002177.1, and zebrafish retinal variant Impdh1a_tvX1 ({"type":"entrez-nucleotide","attrs":{"text":"XM_005159007.4","term_id":"1207134073","term_text":"XM_005159007.4"}}XM_005159007.4). B, conserved N-terminal alpha-helix in blue and gene beginning in pink. C, residues in orange (ordered) are resolved in cryo-EM structures of the human protein (9), remainder of C-terminus in yellow is flexible and not resolved in structures (9). D, GTP-inhibition curves of zebrafish Impdh1a (544) (solid gray line), Impdh1a_tvX1 (solid black line), and nonassembly Y12A protein (dashed black line). Each data point represents a triplicate and the error bars are standard deviation. impdh, inosine monophosphate dehydrogenase; tv, transcript variant. |
|
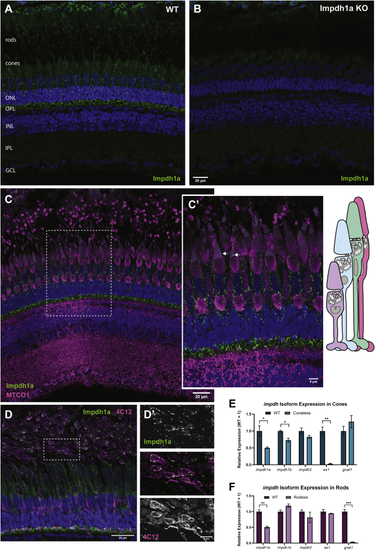
Impdh1a_tvX1 is expressed exclusively in rods and cones.A, representative IHC images showing Impdh1a staining of WT and (B) KO adult zebrafish retina at 11:00 AM using the C-terminal antibody. C, Impdh1a (green) and MTCO1 (magenta) immunostaining of WT-pigmented retina. The nuclei are blue. C′, magnified section of image in (C) showing Impdh1a filament localization in cones. The arrows indicate where Impdh1a filaments appear to form in the OS. Note the tiering of photoreceptors normal in adult zebrafish retina. D, representative Impdh1a staining (green) and 4C12 (magenta) of bleached retina showing the localization of Impdh1a with known zebrafish rod marker. The nuclei are stained blue. D′, magnified section of (D) showing overlap of Impdh1a with 4C12 (rod) staining. E, qPCR quantification of impdh1a, 1b, and 2 transcripts in coneless retina and (F) qPCR analysis of relative impdh1a, 1b, and 2 expression in rodless retina. es1 and gnat1 are cone and rod specific genes, respectively. N = 3 animals and the error bars are standard error. IHC, immunohistochemistry; impdh, inosine monophosphate dehydrogenase. ns p > 0.05, ∗ p ≤ 0.05 , ∗∗ p ≤ 0.01, ∗∗∗ p ≤ 0.001. |
|
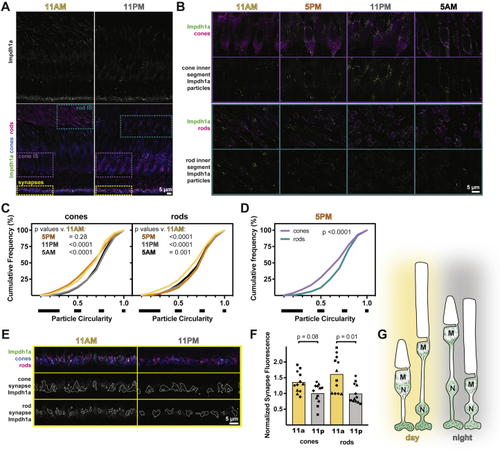
Impdh1a localization in photoreceptors is dynamic.A, representative IHC images of zebrafish outer retina at 11 AM and 11 PM stained with antibodies against Impdh1a (top white and bottom green) and colabeled (bottom panels) with transgenic cone-targeted GFP (blue) and rods (4C12, magenta). B, detailed IHC images of photoreceptor inner segments throughout the day. Impdh1a (green) counterstained with cone or rod markers (magenta). C, cumulative frequency distributions of particle circularity ratios in rods and cones throughout the day. N = 4 animals, three experiments each. D, comparison of particle circularity ratios in cones and rods at 5 PM. E, detailed IHC images of photoreceptor synapses at 11 AM and 11 PM. Top, Impdh1a (green) counterstained with cone (blue) or rod (magenta) markers. Middle and bottom, Impdh1a signal overlapping with cone or rod synapse masks, respectively. F, quantification of mean synapse fluorescence in cones and rods at 11 AM and 11 PM, normalized to 11 PM. Each symbol is a different animal, three experiments for each animal. G, hypothesized model for Impdh1a dynamics throughout the day. In daytime, longer Impdh1a filaments occupy the inner segments, surrounding mitochondrial clusters (M) and nuclei (N). More subtle changes occur at synapses, with slightly reduced Impdh1a immunoreactivity at night. IHC, immunohistochemistry; impdh, inosine monophosphate dehydrogenase. |
|
Loss of impdh1a does not alter the expression of impdh1b or impdh2 and does not cause photoreceptor degeneration.A, schematic of impdh1a KO mutation, a G > T mutation at essential splice site between exon 5 and 6 resulting in a premature stop codon 11 amino acids after exon 5. B, primary sequence of impdh1a_tvX1 compared with impdh1a KO. Blue marks start of retained intron, purple marks premature stop codon in impdh1a KO. qPCR analysis of (C) impdh1a, (D) impdh1b, and (E) impdh2 transcripts at 11:00 AM in WT, HET, and impdh1a KO retinas. N = 6 for WT and heterozygous animals and N = 5 for KO animals. F, representative IHC images of 18 month old impdh1a WT and KO retinas with mitochondria (MTCO1) stained magenta. Cone photoreceptors express eGFP under the cone transducin promotor (TaCP:eGFP) and are stained with eGFP antibody to visualize cones (green). The nuclei are stained in blue. The images are maximum intensity projections of 20 stacks 0.3 μm per step. G, control demonstrating lack of Impdh1a protein at 18 months in impdh1a KO retinas. H, mitochondrial (MTCO1) staining of impdh1a WT and KO retinas showing normal mitochondrial localization and morphology in the KO retina at 18 months. I, cone nuclei quantification and (J) outer nuclear layer thickness (rod nuclei) comparing impdh1a WT and KO zebrafish retinas at 18 months. K, rod- (gnat1) and cone- (es1) specific transcripts in WT compared with KO. impdh2 transcript, which is not found in rod or cones, used as a control. N = 5 animals for WT and N = 3 animals for KO. L, whole retina mtDNA copy number after feeding or after 18 to 24 h fasting; N = 3 for all the samples. M, representative TEM images of 18 month old WT and KO retinas showing high resolution morphology of retinal layers (left and middle panel) and mitochondria (right) in 18 months impdh1a WT and KO retinas. impdh, inosine monophosphate dehydrogenase; tv, transcript variant. ns p > 0.05, ∗ p ≤ 0.05. |
|
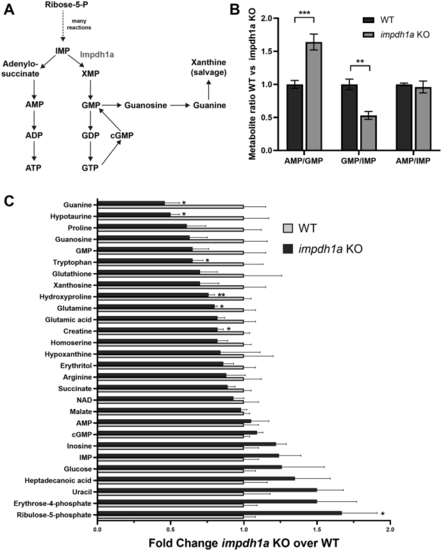
Steady state guanine levels are dramatically reduced in impdh1a KO retinas.A, a schematic of purine nucleotide synthesis pathway. B, the ratio of metabolites directly connected to Impdh enzyme activity. C, quantification from LC/MS/MS analysis of a subset of relevant metabolites and metabolites at 11:00 AM with significant changes between WT and impdh1a KO. The experiment was repeated twice, total N = 5 or 6 animals, and the error bars are standard error. impdh, inosine monophosphate dehydrogenase. ns p > 0.05, ∗ p ≤ 0.05, ∗∗ p ≤ 0.01, ∗∗∗ p ≤ 0.001. |