- Title
-
SOCS3 is Related to Cell Proliferation in Neuronal Tissue: An Integrated Analysis of Bioinformatics and Experiments
- Authors
- Yu, Y., Sung, S.K., Lee, C.H., Ha, M., Kang, J., Kwon, E.J., Kang, J.W., Kim, Y., Kim, G.H., Heo, H.J., Lee, H., Kim, T.W., Lee, Y., Myung, K., Oh, C.K., Kim, Y.H.
- Source
- Full text @ Front Genet
|
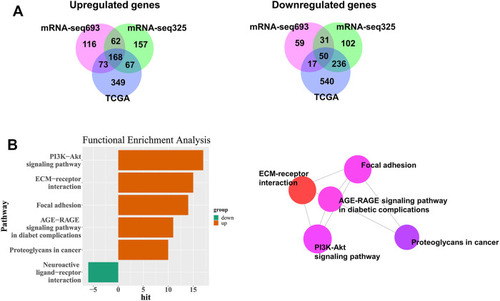
FIGURE 1. Workflow of gene expression analysis of RNA-seq datasets TCGA and CGGA. Each datasets were run through the pipeline individually, then selected common DEGs between GBM and LGG. |
|
Identification of DEGs among TCGA and CGGA datasets of GBM and LGG. |
|
Univariate survival analysis in GBM and LGG stratified by SOCS3 expression based on the TCGA and CGGA data. |
|
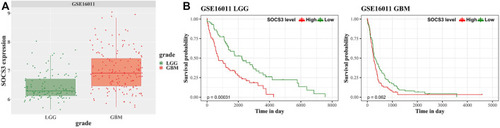
Univariate survival analysis in GBM and LGG stratified by SOCS3 expression based on the GSE16011 dataset for validation. |
|
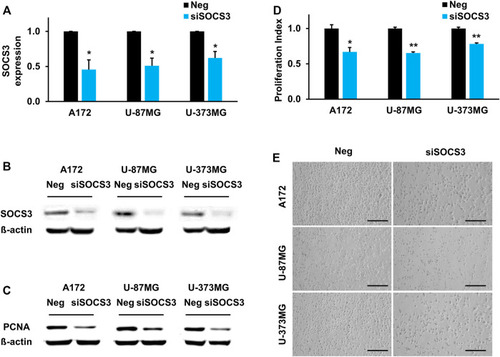
SOCS3 governs brain development by regulating proliferation capacity. |
|
The zebrafish analogue socs3b governs brain development by regulating proliferation. PHENOTYPE:
|