- Title
-
MIC-Drop: A platform for large-scale in vivo CRISPR screens
- Authors
- Parvez, S., Herdman, C., Beerens, M., Chakraborti, K., Harmer, Z.P., Yeh, J.J., MacRae, C.A., Yost, H.J., Peterson, R.T.
- Source
- Full text @ Science
|
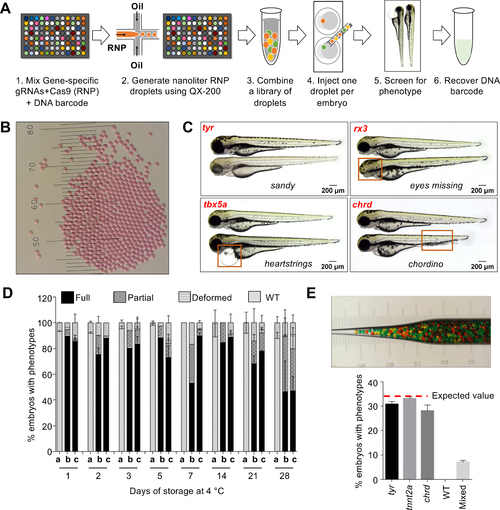
MIC-Drop enables high-throughput CRISPR screens in zebrafish.
(A) Workflow of the MIC-Drop platform. A microfluidics device generates nanoliter-sized droplets, each containing ribonucleoproteins (RNP) targeting a gene-of-interest and a unique DNA barcode associated with the gene. Droplets targeting multiple genes are intermixed, loaded into a single injection needle and injected serially into one-cell zebrafish embryos. Embryos showing phenotypes-of-interest are isolated and the causative genotype is identified by retrieving and sequencing the barcode. (B) Droplets are uniform in size. Distance between bars is 0.1 mm. (C) Injection of droplets containing RNPs targeting tyr, rx3, tbx5a, and chrd genes recapitulates known mutant phenotypes in F0, highlighted by red boxes. (D) RNP-containing droplets are non-toxic and stable for prolonged storage – retaining activity at least 28 days of storage at 4°C. a: Uninjected; b: Traditional RNP injection; c: MIC-Drop injection. N = (Day1: a-118, b-79, c-134; Day2: a-116, b-68, c-59; Day3: a-120, b-88, c-87; Day5: a-129, b-78, c-95; Day7: a-105, b-102, c-107; Day14: a-94, b-53, c-77; Day21: a-123, b-100, c-132; Day28: a-114, b-100, c-94. (E) Single-needle injection of intermixed droplets targeting three different genes (tyr, tnnt2a, chrd) and subsequent phenotyping shows even representation of each droplet with all of the embryos exhibiting only one of the three expected phenotypes, and none (0%) were wildtype. Total N sequenced = 230 from 3 separate injections. Inset: Hundreds of pseudo-colored droplets (used as proxies for droplets targeting different genes) do not fuse when transferred to an injection needle. |
|
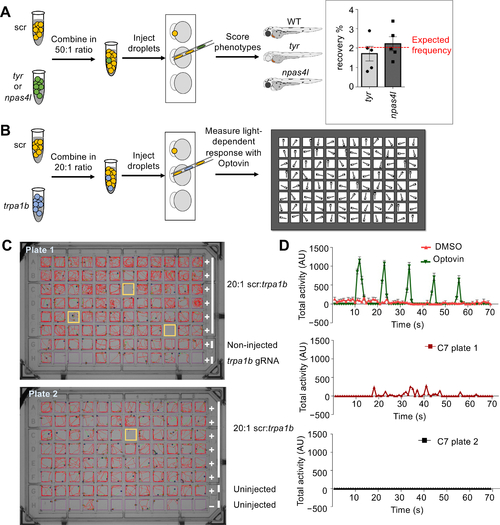
MIC-Drop enables large-scale phenotypic screens and small molecule target identification.
Schematic of a spike-in (A) phenotypic and (B) behavioral screen to test robustness of the MIC-Drop platform. (A) For the phenotypic screen, droplets targeting either tyr or npas4l were intermixed with droplets containing non-targeting scrambled gRNAs (scr) in a 1:50 ratio. After single-needle droplet injection, the percentage of embryos showing albino or cloche phenotypes was scored. Inset: The albino and cloche phenotypes are recovered at a frequency of ~2%, which is the expected frequency from a 1:50 ratio mix. (B) Similar to A, except droplets targeting trpa1b were intermixed with scr droplets in a 1:20 ratio. Following injection, embryos were arrayed in a multi-well plate, treated with optovin, and assayed for light-dependent motor response. (C) Tracking and (D) quantitation of zebrafish movement showed that embryos injected with droplets targeting trpa1b are refractory to optovin- and light-induced motion response. |
|
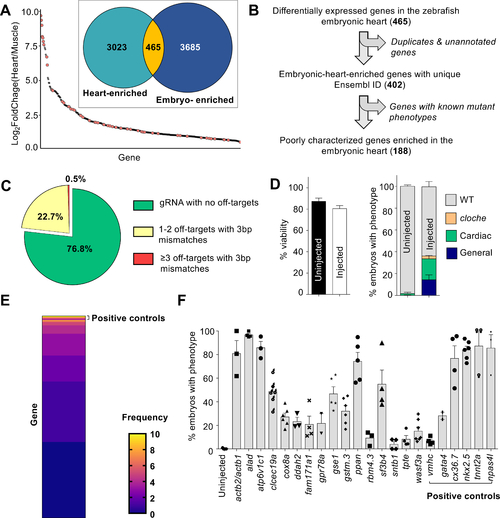
A genetic screen to identify novel regulators of cardiovascular development.
(A) A publicly available dataset was used to populate a list of candidate genes enriched in the embryonic zebrafish heart. ~14% of the genes (orange dots) have reported cardiac phenotypes in ZFIN suggesting enrichment of genes important in heart development. (B) Filtering to remove genes with known mutant phenotypes yields 188 poorly-characterized genes potentially important for cardiovascular development in zebrafish. (C) gRNA sequences with fewer off-targets were prioritized. (D) MIC-Drop screen of the 188 candidate genes and subsequent phenotyping shows no significant differences in viability between uninjected and droplet-injected embryos by 3 dpf N = (Left panel: Uninjected-1801, Injected-2502; Right panel: Uninjected-1571, Injected-2013). Embryos with gross morphological defects at 3 dpf (~15%) were removed and the barcodes of those with cardiac defects were sequenced. Droplets targeting npas4l were spiked-in at 2% proportion as positive control. (E) Barcode sequencing of embryos displaying any cardiac phenotype (e.g. looping defect, chamber dysmorphogenesis, valve defect, arrhythmia, etc.) yields “hit” candidates. Heat map shows the observed frequency of each barcode. As positive controls, barcodes for tnnt2a, nkx2.5, and npas4l are enriched in the set of embryos exhibiting any visible cardiac phenotype. Genes with barcode frequency of ≥ 4 (Binomial probability ≤ 0.05) or with consistent cardiac phenotypes were considered for secondary validation. (F) Secondary validation by direct RNP injection corroborates screening results and identifies a dozen novel genes, the loss of which results in cardiac phenotypes in at least 20% of F0 embryos. N = (WT-238; actb2/actb1-124; alad-137; atp6v1c1-135; clec19a-182; cox8a-130; ddah2-126; fam171a1-259; gpr78a-186; gse1-266; gstm.3-199; ppan-304; rbm4.3-153; sf3b4-307; sntb1-107; tpte-209; wasf3a/b-130; vmhc-107; gata4-106; cx36.7-147; nkx2.5-186; tnnt2a-124; npas4l-103). |
|
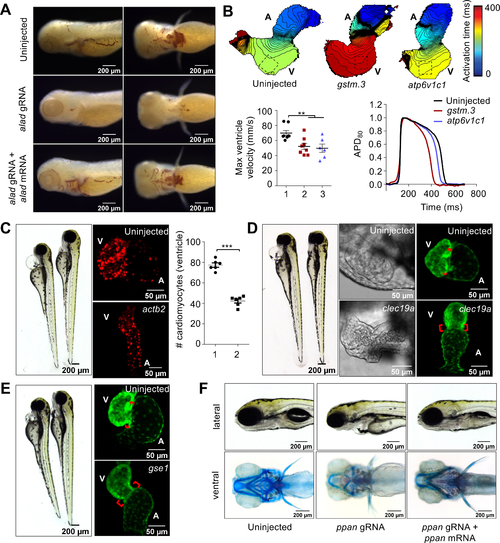
CRISPR screen using MIC-Drop identifies novel genes responsible for cardiovascular development.
(A) o-dianisidine staining shows loss of alad results in porphyria, which can be rescued by co-injection of alad mRNA. (B) Loss of gstm.3 or atp6v1c1 results in abnormal cardiac electrophysiology. Isochronal maps and action potential measurements reveal reduced conduction velocities, and shorter ventricular action potential duration in the gstm.3 and atp6v1c1 crispants relative to uninjected controls 1: Uninjected; 2: gstm.3 crispants; 3. atp6v1c1 crispants. Data are presented as mean ± sem (** = p≤0.01; *** = p≤0.001). Loss of (C) actb2 (D) clec19a (E) gse1, and (F) ppan result in distinct cardiac malformations. actb2 crispants have a small ventricle with reduced number of ventricular cardiomyocytes 1: Control; 2: actb2-targeting gRNAs. (C). Loss of clec19a and gse1 result in abnormal morphogenesis and an extended atrioventricular canal relative to wildtype embryos (D-E). Alcian blue staining of ppan crispants shows abnormal jaw and skull development, which is rescued by ppan mRNA injection. The embryos also display cardiac edema, and a silent ventricle (F). |