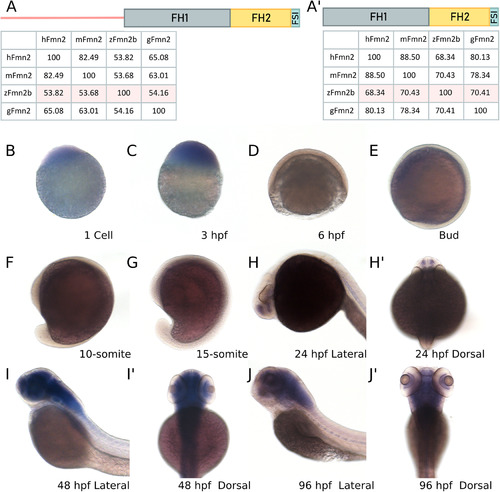
- Title
-
The Formin Fmn2b Is Required for the Development of an Excitatory Interneuron Module in the Zebrafish Acoustic Startle Circuit
- Authors
- Nagar, D., James, T.K., Mishra, R., Guha, S., Burgess, S.M., Ghose, A.
- Source
- Full text @ eNeuro
|
|
|
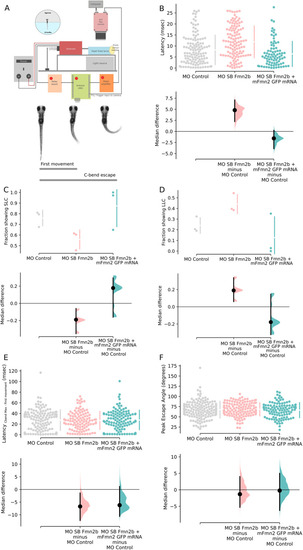
Fmn2b knock-down increased the latency to respond to mechano-acoustic stimulus. Fmn2b knock-down using translation and splice blocking morpholinos (morpholino validation in Extended Data PHENOTYPE:
|
|
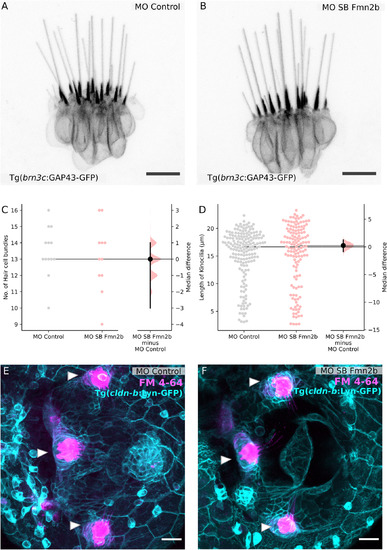
Sensory components of the acoustic startle circuit are not affected in Fmn2b morphants. Hair cells of the lateral crista of the zebrafish inner ear are visualized using the Tg ( |
|
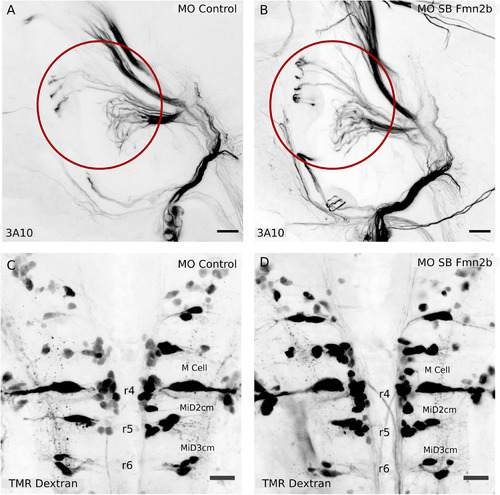
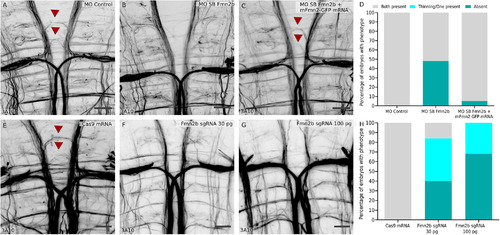
The SAG relaying sensory information and the hindbrain reticulospinal neurons are not affected in Fmn2b morphants. Whole-mount immunostaining with anti-neurofilament antibody 3A10 was used to visualize the SAG connecting to the M-cells in 96-hpf larvae with ( |
|
Fmn2b knock-down impairs axonal outgrowth in the spiral fiber tracts. Whole-mount immunostaining using anti-neurofilament antibody 3A10 stains axons in 96-hpf zebrafish hindbrain ( |