- Title
-
Candida auris Cell Wall Mannosylation Contributes to Neutrophil Evasion through Pathways Divergent from Candida albicans and Candida glabrata
- Authors
- Horton, M.V., Johnson, C.J., Zarnowski, R., Andes, B.D., Schoen, T.J., Kernien, J.F., Lowman, D., Kruppa, M.D., Ma, Z., Williams, D.L., Huttenlocher, A., Nett, J.E.
- Source
- Full text @ mSphere
|
|
|
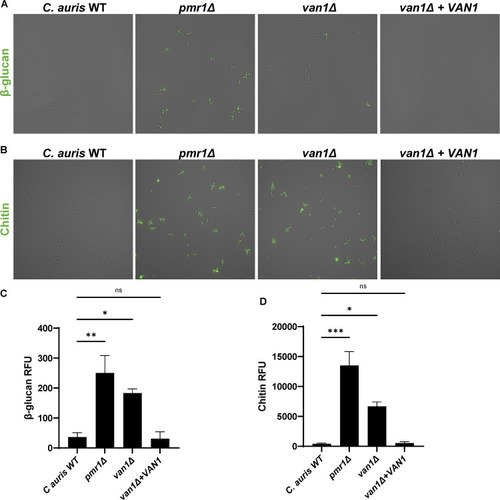
C. auris mannosylation pathway mutants are susceptible to neutrophil attack. (A) C. auris strains were incubated with human neutrophils for 1 h and were subsequently imaged via scanning electron microscopy. Images are 10,000× magnification, measurement bars represent 1 μm. (B and C) Human neutrophils were labeled with calcein-AM (green) and cocultured with individual C. auris strains labeled with calcofluor white (blue) for 1 h and imaged via fluorescence microscopy (B). The numbers of neutrophils engulfing fungal cells were counted and the percentages of total engaged neutrophils were calculated (C); n ≥ 3, mean with standard error of the mean (SEM) shown. (D) Individual C. auris strains were cultured with human neutrophils for 4 h and viable burden was estimated by PrestoBlue metabolic activity following neutrophil lysis; n = 3, mean with standard deviation shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant by one-way ANOVA with Holm-Sidak multiple comparisons to C. auris WT. |
|
|
|
|
|
|
|
|
|
|