- Title
-
Exocrine pancreas proteases regulate β-cell proliferation in zebrafish ciliopathy models and in murine systems
- Authors
- Hostelley, T.L., Nesmith, J.E., Larkin, E., Jones, A., Boyes, D., Leitch, C.C., Fontaine, M., Zaghloul, N.A.
- Source
- Full text @ Biol. Open
|
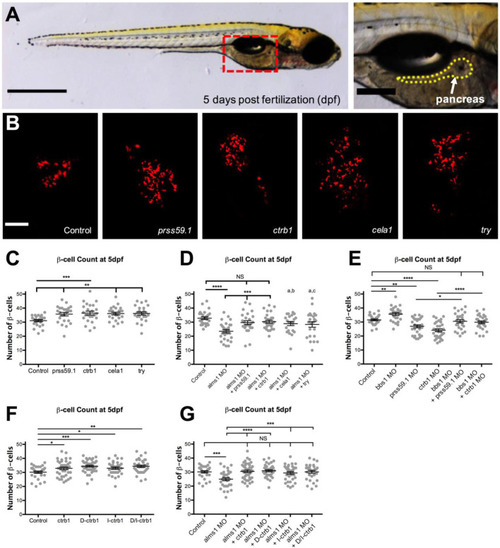
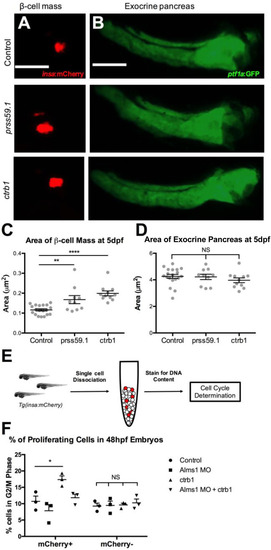
PHENOTYPE:
|
|
|
|
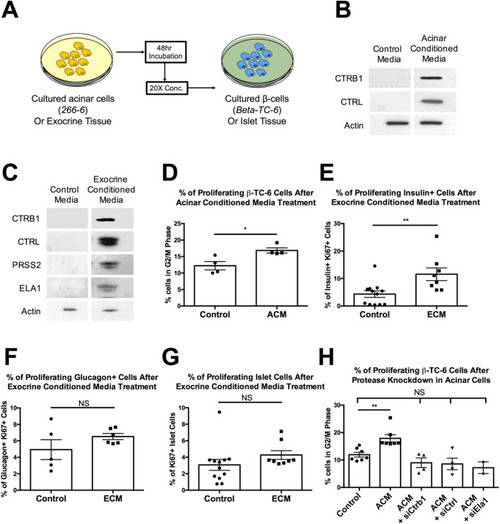
(A) Schematic of experimental design for assessing protease-β-cell interaction in cultured β-cells and islets. (B) Western blot of β-TC-6 lysate from cells treated with control media or acinar conditioned media (ACM) using anti-CTRB1 and anti-CTRL antibodies. (C) Western blot of islet tissue lysate from islets treated with control media or exocrine conditioned media (ECM) using anti-CTRB1, anti-CTRL, anti-PRSS2 and anti-ELA1 antibodies. (D) Quantification of proliferating β-TC-6 cells by percentage of cells in G2/M phase in control media or ACM treated cells. (E) Quantification of proliferating β-cells (insulin+) by percentage of Ki67 positive cells in control media or ECM treated islets. (F) Quantification of proliferating α-cells (glucagon+) by percentage of Ki67 positive cells in control media or ECM treated islets. (G) Quantification of proliferating islet cells by percentage of Ki67 positive cells in control media or ECM treated islets. (H) Quantification of proliferating β-TC-6 cells by percentage of cells in G2/M phase in control media, ACM, ACM lacking CTRB1 (ACM+siCtrb1), ACM lacking CTRL (ACM+siCtrl) and ACM lacking ELA1 (ACM+siEla1) treated cells. (**=P<0.01, Ordinary one-way ANOVA). Error bars represent standard error of the mean, symbols represent the following significance: NS=P>0.05, *=P<0.05, **=P<0.01, ***=P<0.001, Student's t-test unless otherwise indicated. |
|
|
|
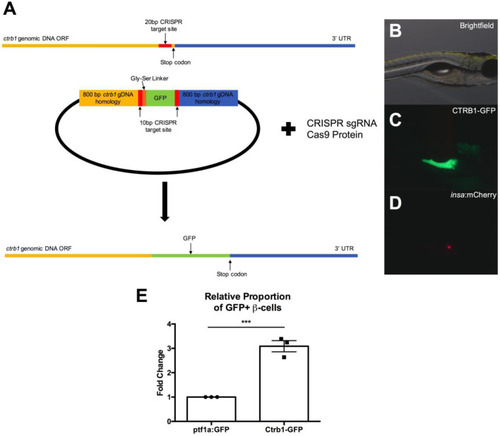
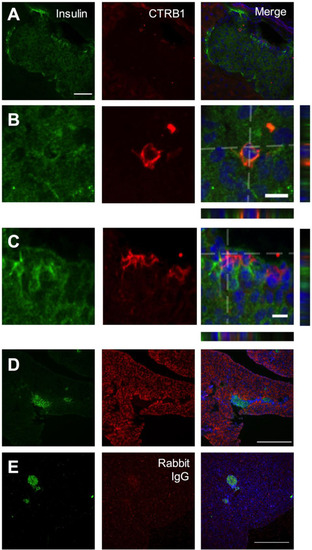
(A) High magnification islet view. Scale bar: 50 μm. (B,C) High magnification β-cell view. Merged image is a compressed Z-stack, orthogonal XZ and YZ are provided. Scale bars: 10 μm. (D,E) Low magnification pancreas stack, orthogonal XZ and YZ are provided. Scale bars: 10 μm. (D,E) Low magnification pancreas. |