- Title
-
Wnt3 Is Lipidated at Conserved Cysteine and Serine Residues in Zebrafish Neural Tissue
- Authors
- Dhasmana, D., Veerapathiran, S., Azbazdar, Y., Nelanuthala, A.V.S., Teh, C., Ozhan, G., Wohland, T.
- Source
- Full text @ Front Cell Dev Biol
|
Schematic for a SPIM-FCS experiment. |
|
Influence of C80 and S212 lipidation on secretion |
|
Secretion of positive and negative control in the BV |
|
Influence of C80 and S212 lipidation on membrane localization |
|
Expression profile of the mutants co expressed with PMT-mApple. Co-expression of mutants (Wnt3C80A-EGFP, Wnt3S212A-EGFP, and Wnt3S212AC80A-EGFP) and PMT-mApple in the zebrafish brain at ∼48 hpf. PMT-mApple is a protein located on the inner leaflet of the cell membrane. It is expressed under a 4kb Wnt3 promoter and serves to mark the source cells of Wnt3 expression. |
|
Membrane localization and dynamics of PMT-mEGFP Cell membrane localization of PMT-mEGFP (left) and a representative autocorrelation function (ACF; dots) and fit (line) of FCS measurement at the cell membrane (right). The FCS curves were fitted using 2D-2particle-1triplet model. Scale bar 10 μm. |
|
Representative SPIM-FCS and Diffusion law data from a cell. |
|
Influence of C80 and S212 lipidation on the interaction of Wnt3 with Fzd1 receptor. |
|
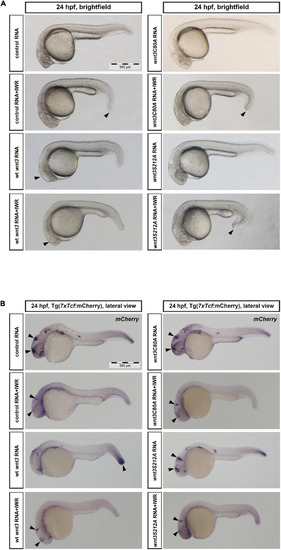
Influence of C80 and S212 residues on Wnt signaling activity |
|
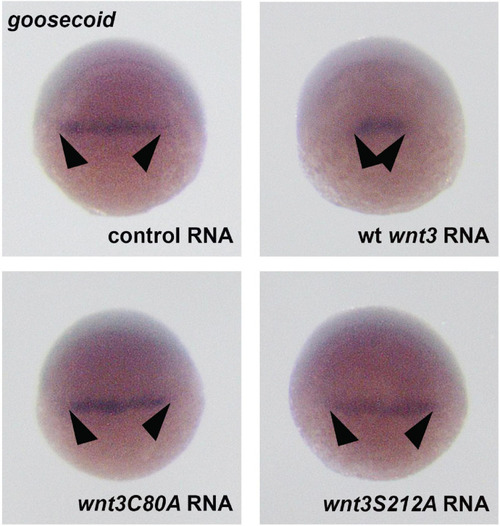
Whole mount in situ hybridization for the dorsal organizer marker gene goosecoid (gsc) in wt embryos at the shield stage, showing the effects of control (EGFP 100 pg, 35/35 embryos, arrowheads: normal gsc expression domain), Wnt3-EGFP (100 pg, 31/35 embryos, arrowheads: reduction in gsc expression domain), Wnt3C80A EGFP (100 pg, 38/41 embryos, arrowheads: no effect on gsc expression domain), and Wnt3S212A-EGFP (100 pg, 32/34 embryos, arrowheads: no effect on gsc expression domain). |