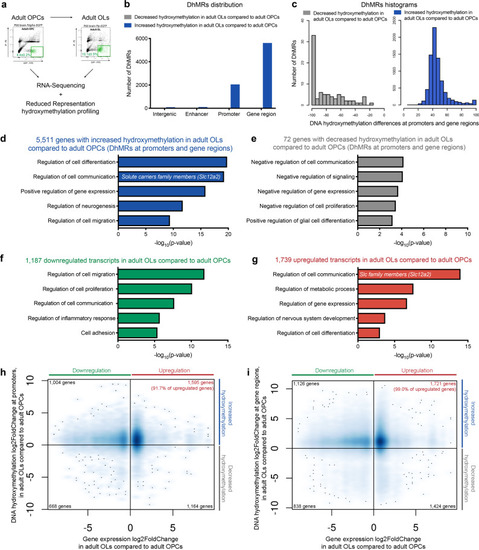
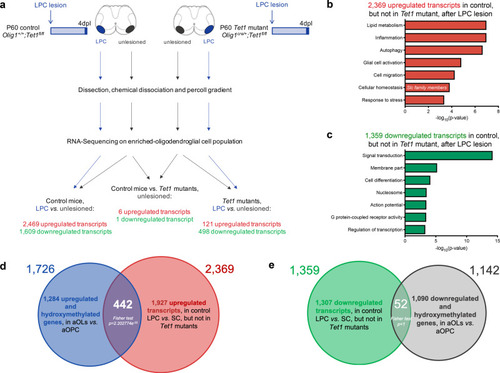
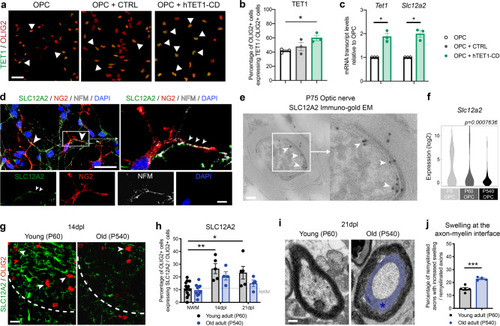
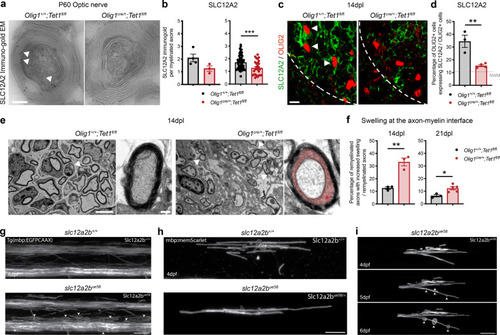
|
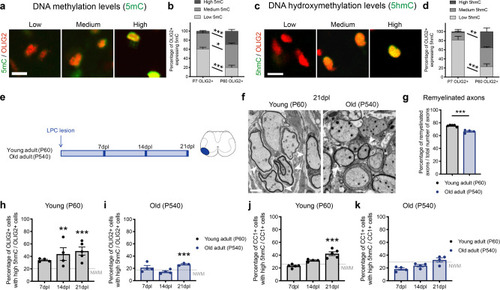
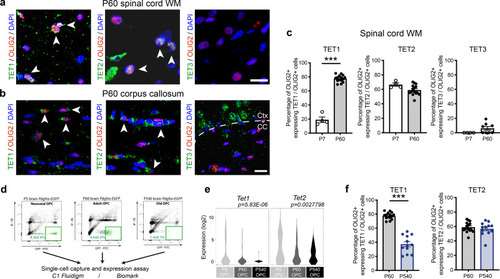
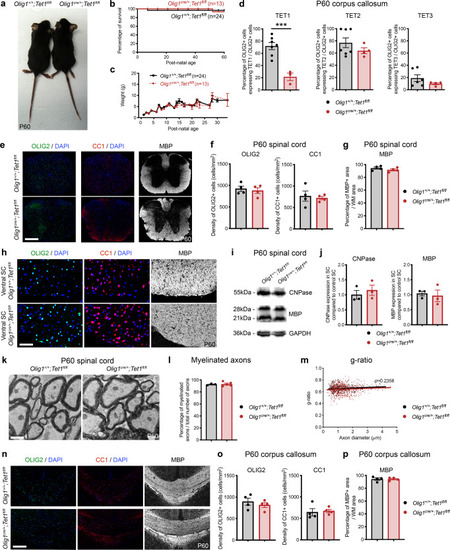
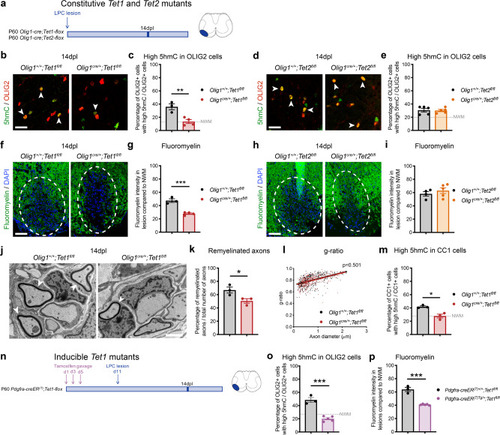
Ablation of <italic>Tet1</italic> during development is compatible with normal myelination in the adult central nervous system.a Photograph of adult controls (Olig1+/+;Tet1fl/fl) and Tet1 mutant (Olig1cre/+;Tet1fl/fl) mice reveals no difference in body size. b Kaplan–Meier survival curve and c body weight for controls (n = 24) and Tet1 mutants (n = 13). Error bars represent SEM. d Percentage of TET1, TET2, and TET3 expressing OLIG2 cells in the corpus callosum of mice of the two genotypes. Note decreased percentage of OLIG2 + cells expressing TET1 without compensatory increase of TET2 or TET3 expressing cells. Error bars represent SEM for n = 7 control mice and n = 4 Tet1 mutants (***p = 0.0010 for TET1, p = 0.2801 for TET2, p = 0.2338 for TET3, Student’s t test two-tailed). e Representative P60 coronal sections of Olig1+/+;Tet1fl/fl and Olig1cre/+;Tet1fl/fl spinal cords, stained for OLIG2 (green), CC1 (red), MBP (white), DAPI (blue). Scale bars = 500 µm. f Quantification of OLIG2 + and CC1 + cell density (number of cells per mm2) in the spinal cord of mice of the indicated genotypes. Error bars represent SEM for n = 4 mice (p = 0.6482 for OLIG2, p = 0.7252 for CC1, Student’s t test two-tailed). g Average MBP + area relative to total white matter (WM) area in coronal spinal cord sections. Error bars represent SEM for n = 4 mice (p = 0.2693, Student’s t test two-tailed). h Representative confocal micrograph of adult ventral spinal cord in coronal sections from mice of the indicated genotype, stained for OLIG2 (green), CC1 (red), MBP (white), DAPI (blue). Scale bars = 100 µm. i Representative Western blot of protein extract from control and Tet1 mutants reveals similar levels of the myelin proteins CNPase, and MBP. GAPDH used as loading control. Molecular weight indicated on the left. j Average levels of the myelin proteins CNPase (left) and MBP (right), relative to GAPDH, quantified in three independent experiments. Error bars represent SEM for n = 3 spinal cords (p = 0.5347 for CNPase, p = 0.7295 for MBP, Student’s t test two-tailed). k Representative ultra-micrograph of adult ventral white matter spinal cords from mice of the indicated genotype revealing no difference in myelination between controls and mutants. Scale bar = 5 µm. l Percentage of myelinated axons relative to total axons in sections of P60 Olig1+/+;Tet1fl/fl and Olig1cre/+;Tet1fl/fl white matter spinal cords. Error bars represent SEM for n = 3 control mice and n = 5 Tet1 mutants (p = 0.8009, Student’s t test two-tailed). m Scatter plot of the g-ratio in myelinated axons of the indicated caliber, in adult control (black) and Tet1 mutant (red) spinal cords. A total of n = 116 axons in 3 control mice and n = 199 axons in 5 Tet1 mutants were quantified (non-linear regression, p = 0.2358, comparison of slopes with sum-of-squares F Test). n Representative confocal image of P60 coronal sections from Olig1+/+;Tet1fl/fl and Olig1cre/+;Tet1fl/fl corpus callosum, stained for OLIG2 (green), CC1 (red), MBP (white), DAPI (blue). Scale bar = 500 µm. o Quantification of OLIG2 + and CC1 + cell density (number of cells per mm2) in the corpus callosum of mice of the indicated genotypes. Error bars represent SEM for n = 4 mice (p = 0.4644 for OLIG2, p = 0.7750 for CC1, Student’s t test two-tailed). p Average MBP + area relative to total white matter (WM) area in corpus callosum. Error bars represent SEM for n = 4 mice (p = 0.9960, Student’s t test two-tailed).
|