- Title
-
A forward genetic screen identifies Dolk as a regulator of startle magnitude through the potassium channel subunit Kv1.1
- Authors
- Meserve, J.H., Nelson, J.C., Marsden, K.C., Hsu, J., Echeverry, F.A., Jain, R.A., Wolman, M.A., Pereda, A.E., Granato, M.
- Source
- Full text @ PLoS Genet.
|
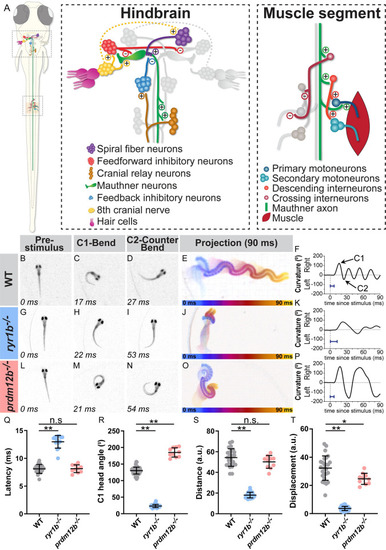
The zebrafish larval startle response is amenable to circuit and genetic analysis. PHENOTYPE:
|
|
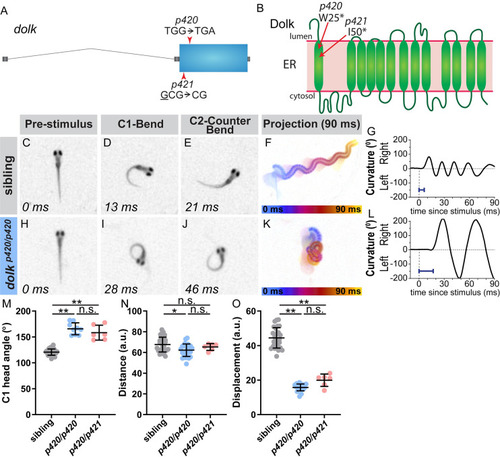
The glycosylation pathway enzyme Dolk regulates the magnitude of the startle response. (A) Gene structure for PHENOTYPE:
|
|
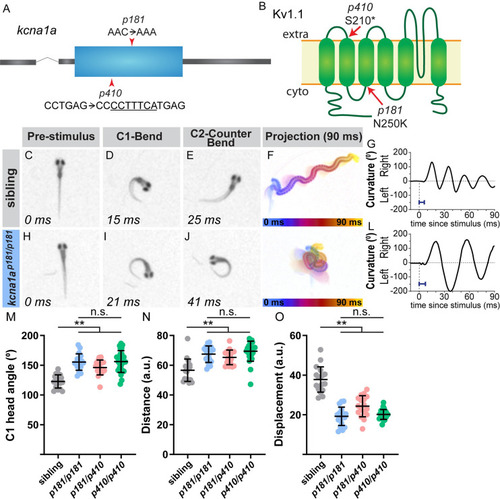
The potassium channel subunit Kv1.1 regulates the magnitude of the startle response. (A) Gene structure for PHENOTYPE:
|
|
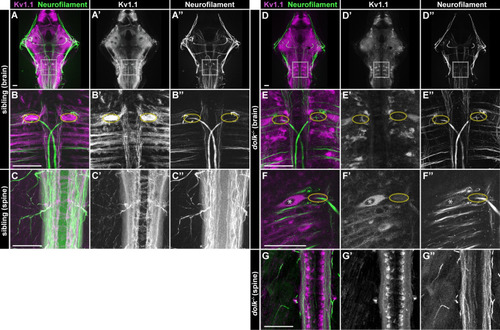
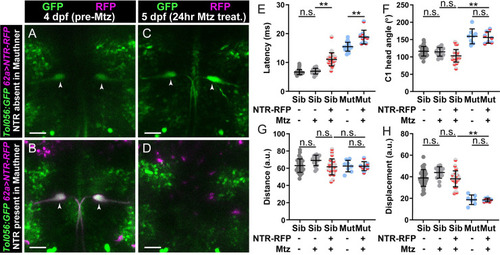
Dolk is required for proper localization of Kv1.1. (A-C) In 5 dpf |
|
Kv1.1 functions outside the Mauthner command neuron to control swim movement magnitude. (A-D) |
|
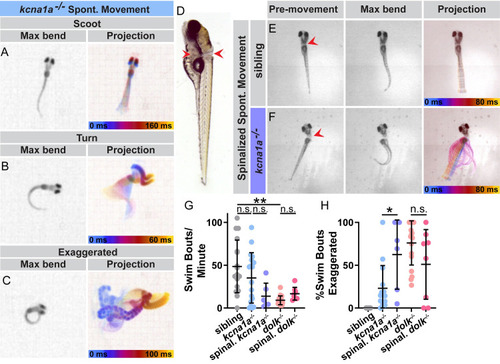
Kv1.1 and Dolk function in the spinal cord to control movement magnitude. (A-C) Examples of spontaneous swim movements performed by PHENOTYPE:
|
|
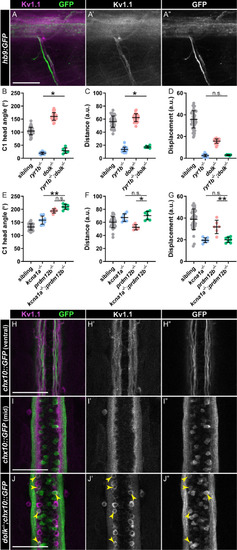
Kv1.1 is expressed in motor neurons and V2a interneurons. (A) Kv1.1 (magenta:A; grey:A’) localizes to motor axons expressing hb9:GFP (green:A; grey:A”). (B-G) Kinematic parameters of the acoustic startle response in (B-D) sibling, |