- Title
-
A duplication on chromosome 16q12 affecting the IRXB gene cluster is associated with autosomal dominant cone dystrophy with early tritanopic color vision defect
- Authors
- Kohl, S., Llavona, P., Sauer, A., Reuter, P., Weisschuh, N., Kempf, M., Dehmelt, F.A., Arrenberg, A.B., Sliesoraityte, I., Zrenner, E., van Schooneveld, M.J., Rudolph, G., Kühlewein, L., Wissinger, B.
- Source
- Full text @ Hum. Mol. Genet.
|
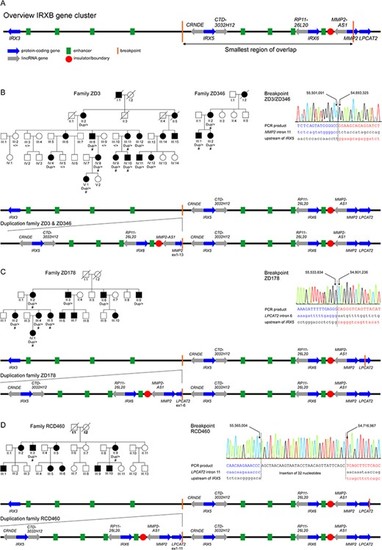
Tandem duplications at the IRXB gene cluster on chromosome 16q12 segregate with autosomal dominant cone dystrophy in four families. (A) Genomic organization of the IRXB gene cluster and the other two genes involved in the duplications, MMP2 and LPCAT2. Blue arrows indicate the protein-coding genes IRX3, IRX5, IRX6, MMP2 and LPCAT2, and gray arrows the non-coding RNAs CRNDE, CTD-3032H12, RP11-26 L20 and MMP2-AS1. The IRXB locus is highly conserved throughout evolution, not only in its genes but also its regulatory elements. Conserved enhancer elements are depicted in green and insulators/boundaries are represented with a red circle (4). The smallest region of overlap of the duplications found in this study is depicted as by vertical orange lines. (B–D) Pedigrees and segregating genotypes for the analyzed subjects and families in this study, breakpoint mapping and genomic organization of the duplication. Pedigrees: Patients for which DNA was available for CNV analysis are indicated by the identified genotype beneath the individual identifier. The presence of the duplication is indicated by ``dup'', while ``+'' indicates normal wild-type alleles. Patients for which clinical data was available are presented in Figure 3, Table 2 and Supplementary Material, Table S1 and are indicated by ``#''. Circles depict females and squares males, affected individuals by filled and unaffected individuals by open symbols. Already deceased individuals are marked by a diagonal slash. Breakpoint mapping in the investigated families: Electropherograms of the breakpoint sequences as obtained from Sanger sequencing of PCR products covering the breakpoints. Left and right junction sequences are given in blue and red, respectively. The reference sequences are given in lowercase letters below the sequence for the PCR product. Genomic positions refer to GRCh38/hg38. Schematic representation of the tandem repeated duplication as observed in the families in this study in comparison to the genomic organization of the wild-type IRXB gene cluster in families ZD3 and ZD346 (B), family ZD178 (C) and family RCD460 (D). The tandem repeated sequence is expected to create a new topological associated domain (TAD) flanked by the insulators as boundaries. |
|
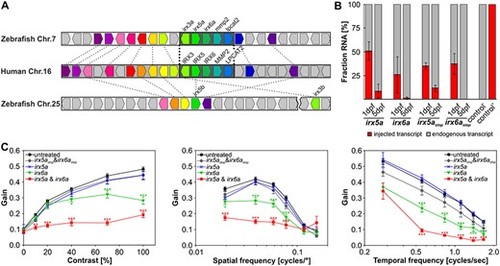
Overexpression of Irx6a and Irx6a & Irx5a cause reduced optokinetic reflex response in zebrafish larvae. (A) Gene synteny between the human chromosome 16 locus containing the IRXB cluster and the zebrafish chromosomes 7 and 25. Using the Genomicus database (http://www.genomicus.biologie.ens.fr/genomicus-97.01/cgi-bin/search.pl) and the UCSC genome browser (http://genome-euro.ucsc.edu/index.html), the conservation of the IRXB cluster between human and zebrafish was analyzed. The schematic comparative gene organization is shown and homologous genes are coded by the same color and are connected by dashed lines between the two analyzed species, whereas nonhomologous genes are depicted in gray. There is a high degree of synteny between human chromosome 16 and zebrafish chromosome 7, whereas the overall synteny between human chromosome 16 and zebrafish chromosome 25 is low for the analyzed 25 human chromosome 16 genes within and flanking the IRXB cluster. Strikingly, the gene cluster spanning IRX3, IRX5, IRX6, MMP2 and LPCAT2 is completely conserved between human and zebrafish. The genes irx5 and irx3 are duplicated in the zebrafish genome with the ‘a’-paralogs being located on chromosome 7 and the ‘b’-paralogs on chromosome 25—the distance between irx5b and irx3b is around 340 kb (GRCz11/danRer11) as depicted by the break on chromosome 25. irx6a, mmp2 or lpcat2 are not duplicated in the zebrafish genome. Thus, the zebrafish genome possesses one highly conserved IRXB cluster on chromosome 7. (B) Relative comparison between endogenously expressed irx5a and ixr6a versus injected transcripts (50 pg cRNA per oocyte) in 1 and 5 dpf old zebrafish embryos/larvae using pyrosequencing. In 1 dpf embryos, we detected an allelic ratio compatible with four copies of irx5a or three copies of irx6a. In 5 dpf larvae, the injected irx5a and irx6a transcripts were barely detectable. A second set of transcripts carrying a premature stop codon (irx5astop and irx6astop) was injected (50 pg cRNA per oocyte) and was detected in similar amounts compared to the injected irx5a and ixr6a transcripts at both time points analyzed. The irx5astop and irx6astop served later on as controls for the functional assay (OKR). RNA of untreated larvae (control −) or plasmid DNA containing a variant for pyrosequencing detection (control +) was used as controls for pyrosequencing. Data are presented as mean ± SEM. (C) Optokinetic response (OKR) analysis with respect to contrast sensitivity and spatial and temporal frequency measured in 5 dpf old treated larvae in comparison to untreated controls. Overexpression of Irx5a did not affect OKR for any of the analyzed stimulus conditions in 5 dpf larvae. Overexpression of Irx6a (equivalent to approximately three gene copies at 1 dpf) induced significant OKR alteration in comparison to untreated larvae at high contrasts and at a variety of spatial and temporal frequencies. Simultaneous overexpression of both Irx6a and Irx5a resulted in an even more reduced OKR when comparing to overexpression of Irx6a alone. Additionally, irx5a and ixr6a transcripts harboring a premature stop codon (irx5astop and irx6astop) were injected to verify that the reduced OKR in irx6a and irx6a & irx5a larvae is indeed due to Irx overexpression. OKR with respect to contrast sensitivity and spatial frequency measured in 5 dpf old larvae treated with 50 pg irx5astop or irx6astop did not show any effect for different contrast or spatial stimuli in comparison to untreated controls. For the tested temporal frequencies, reduced OKR responses were detected at high frequencies. For clarity, statistical significance of only treated versus untreated samples are depicted. Two-way ANOVA with Bonferroni multiple comparison was performed and the results of the statistical test within all groups are summarized in Supplementary Material, Tables S4–S7. Data are presented as mean ± SEM. Nuntreated = 60 larvae. Ntreated = 24 larvae per group. P ≤ 0.001 is indicated with ***, P ≤ 0.01 with ** and P ≤ 0.05 is displayed with *. |
|
Clinical presentation of patients affected by autosomal dominant cone dystrophy caused by duplications at the IRXB gene cluster: (A) Fundus autofluorescence (FAF) and optical coherence tomography (OCT) images of 13 patients with a duplication at the IRXB gene cluster, sorted by the age at examination (compare Fig. 1, Table 2 and Supplementary Material, Table S1). Note the central hypoautofluorescence and paracentral hyperautofluorescence on FAF imaging in the younger patients and the well-demarcated central hypoautofluorescence in the older patients, as well as the central thinning of the outer nuclear layer, the paracentral hyporeflectivity of the retinal pigment epithelium, and/or a subretinal cleft on OCT imaging in the younger patients and the central atrophy in the older patients. (B) Electroretinography (ERG) of six patients sorted by age, commencing with the youngest in light gray and ending with the oldest in black. The normative data are highlighted in gray bars. Under scotopic light conditions, the ERG (indicated by ‘DA’, left) shows some decline, which may be explained by an ageing effect. The photopic ERG (indicated by ‘LA’, middle: single flash; right: 30 Hz flicker) shows a decline in the amplitude of the a- and b-wave. |