- Title
-
Delay in primordial germ cell migration in adamts9 knockout zebrafish
- Authors
- Carver, J.J., He, Y., Zhu, Y.
- Source
- Full text @ Sci. Rep.
|
Conserved structure of Adamts9 from |
|
Strong |
|
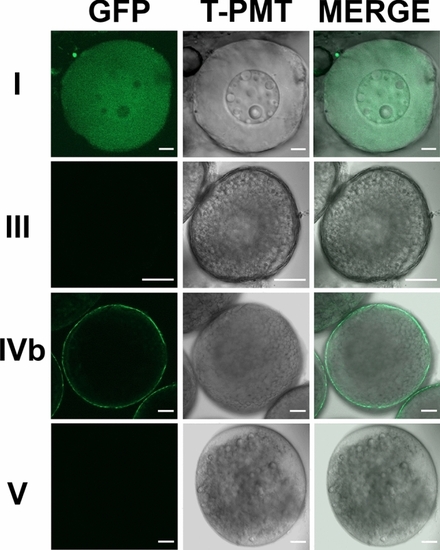
Strong expression of Adamts9 in preovulatory follicles, weak or no expression in immature ovarian follicles or ovulated oocytes in Adamts9 transgenic lines ( |
|
Expression of Adamts9 in F2 transgenic ( |
|
Strong Adamts9 expression in F2 transgenic ( |
|
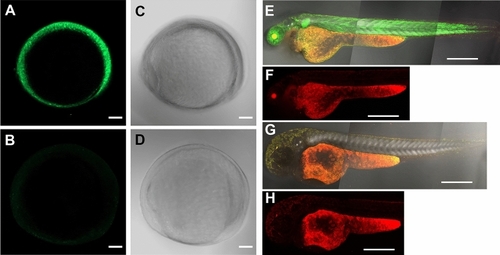
Delayed primordial germ cell (PGC) migration in Adamts9 knockout zebrafish during early development. PGC is labeled with GFP by crossing Adamts9 KO with Tg(vasa:GFP). Distance between two most distant PGCs were determined as an indicator of PGC migration (See Fig. 7 for detail). Showing distance, number of PGC, or representative images of zebrafish embryos at 15, 24 or 48 h post fertilization (hpf) in wildtype (+/+), heterozygous (+/–), and homozygous (–/–) Adamts9 KO. Embryos from at least 4 sets of parents were analyzed. The numbers on the left side of the forward slash is the number of embryos analyzed, and the numbers on the right side of the forward slash indicate sets of parents used for producing these embryos. Top panels are representative confocal images showing entire or part of embryos with GFP labeled PGC at a low magnification (scale bar: 200 μm). Bottom row are magnified confocal images of the embryos from the top (scale bar: 50 μm). |
|
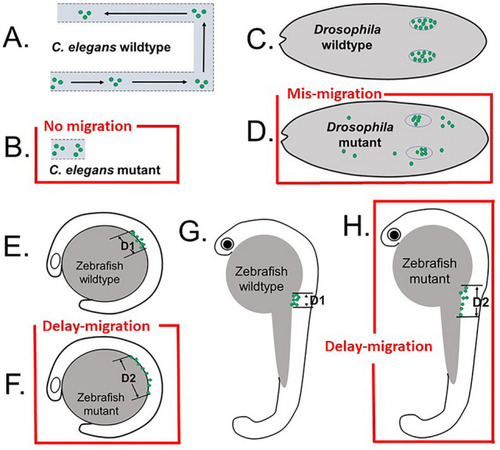
Schematic drawings show difference in the defects of germ cell migration in Adamts9 or its ortholog knockouts in |