- Title
-
Construction of Whole Genomes from Scaffolds Using Single Cell Strand-Seq Data
- Authors
- Hills, M., Falconer, E., O'Neill, K., Sanders, A.D., Howe, K., Guryev, V., Lansdorp, P.M.
- Source
- Full text @ Int. J. Mol. Sci.
|
The principle and some applications of Strand-seq. (A) Strand-seq involves sequencing template strands. Parental homologues (pink and blue) are double stranded; Crick (C) strand in blue, Watson (W) strand in orange. DNA replication occurs in the presence of BrdU, which incorporates into the replicated strand (dotted lines). Sequencing libraries from single daughter cells have BrdU-containing strand selectively removed to generate directional chromosomes; either CC, WW (top) or WC (bottom) depending on segregation. Histograms of directional reads are plotted on ideograms for each chromosome. (B) When homologues inherit different template strands, haplotypes can be determined. In the example, all C reads map to the maternal homologue so all single nucleotide variants (SNVs) identified (black dots) form the maternal haplotype, and all W reads map to the paternal homologue, so all SNVs identified (white dots) form the paternal haplotype. (C) Structural variation can be identified in Strand-seq libraries. Inversions will align to the opposite strand of the reference assembly and as so be identified as a change in template strand state (D) Strand-seq can be used to create assemblies since contigs from the same chromosome will have the same template inheritance pattern. Grouping based on shared template inheritance patterns determines which fragments belong together. Note in the example contigs from ch1, chr3, and chr5 have the same template pattern (WC) so require additional libraries to establish which contigs belong to which chromosome. |
|
of three template inheritance states: WW, WC, or CC (W = blue, C = orange). Through analysis of the template inheritance pattern of multiple cells, scaffolds from the same chromosome share the same pattern and can be resolved. For example, in Cell 1, three chromosomes are represented in linkage group 1 (LG1), but are resolved in subsequent cells. (B) Subsetted data showing 1799 unsorted ferret scaffolds belonging to six linkage groups across 100 cells (CC = blue, WW = orange, WC = grey, no data = white). Prior to clustering (left plot), scaffolds from the same chromosome are unknown, while after clustering (right plot), scaffolds that share template inheritance patterns across individual cells are resolved. Vertical color bar represents called members for each of the six linkage groups. |
|
The effect of different assembly errors or structural variation on clustering. Different errors will generate characteristic patterns in the clustering data. Consider two scaffolds in close proximity on a chromosome, scaffold_1 and scaffold_2. (A) In a case where both scaffolds are oriented in the same direction, the scaffolds will have the same strand-state patterns. When comparing homozygous patterns (WW scaffolds against CC scaffolds), heterozygous patterns (WW or CC scaffolds against WC scaffolds) or comparing all three strand states against each other, there will be high similarity. (B) In the case of a misorientation (or a homozygous inversion), the strand-state patterns will be antithetical when comparing homozygous states, as whenever scaffold_1 is WW, scaffold_2 will be CC, and as such, these scaffolds will be completely dissimilar. However, since misorientations are not visualized in heterozygous inheritance patterns, when comparing WW or CC states against WC states, the scaffolds are highly similar. When comparing all three states against each other, the similarity seen with WC scaffolds and dissimilarity seen with WW or CC scaffolds will cancel out, resulting in ~50% similarity. (C) In cases of a heterozygous inversion, either scaffold_1 or scaffold_2 may have a homozygous state, but not both. Therefore, no comparisons can be made when only considering the homozygous states, and NA values are generated. There will, however, be a high degree of dissimilarity when comparing homozygous and heterozygous states. It is important to distinguish these natural structural variants from assembly reference errors. (D) In cases where a scaffold is incorrectly located to a chromosome (i.e., a chimera), the inheritance pattern between the two scaffolds will be random, and there will be no significant similarity or dissimilarity between these scaffolds. |
|
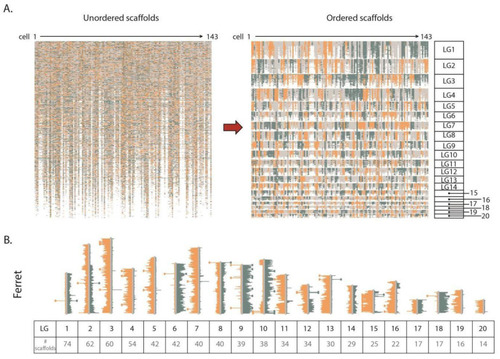
Assemblies made from non-contiguous scaffolds based on Strand-seq data. (A) Left panel shows ferret scaffolds presented in the current assembly order. Orange, blue, and grey represent scaffolds with WW, CC, and WC reads respectively. Right panel shows scaffolds after contiBAIT reordering. (B) Representative ideogram plot of a ferret library after clustering and ordering scaffolds. Each linkage group is represented by a certain number of scaffolds. Chromosomes with WW, WC, and CC inheritance patterns are observed in this library. Changes in strand state represent sister chromatid exchange (SCE) events and are used to map the relative locations of scaffolds. |
|
Assembly misorientations and chimeras are prevalent in early-stage genomes. ( |