- Title
-
Efficient CRISPR-Cas9-Mediated Knock-In of Composite Tags in Zebrafish Using Long ssDNA as a Donor
- Authors
- Ranawakage, D.C., Okada, K., Sugio, K., Kawaguchi, Y., Kuninobu-Bonkohara, Y., Takada, T., Kamachi, Y.
- Source
- Full text @ Front Cell Dev Biol
|
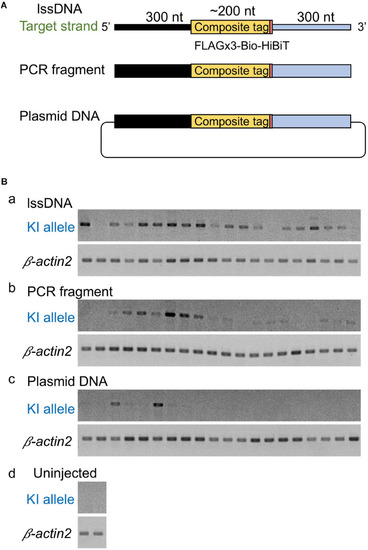
Design of composite tag knock-in through the CRISPR-Cas9 system. In this design, an ∼200-nt length composite that contains either a FLAG or PA epitope tag in its trimeric form followed by a tobacco etch virus (TEV) protease cleavage site, a biotin acceptor domain (Bio tag), and a C-terminus HiBiT peptide tag was knocked into the 3′ end of the coding sequence of the |
|
Cleavage efficiency of the selected crRNA. |
|
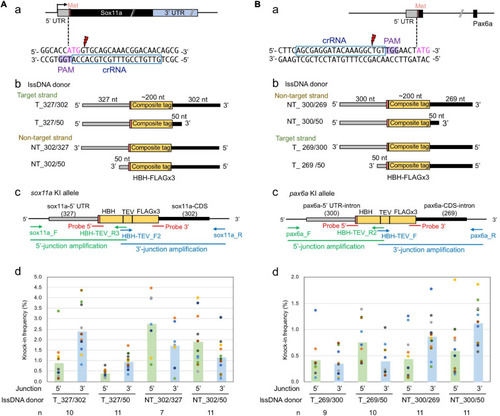
Effect of the lssDNA strand choice on knock-in efficiency. |
|
Comparison of donor DNA template types. |
|
Effect of lssDNA 3′ homology arm length on knock-in efficiency. |
|
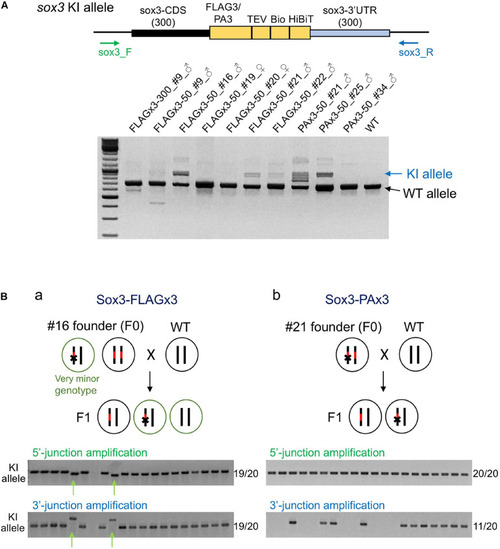
Founder germ cell mosaicism. |
|
Expression of the tagged Sox3 protein. |
|
Effect of lssDNA strand choice and 3′ homology arm length on composite tag knock-in into the |