- Title
-
A biallelic variant in CLRN2 causes non-syndromic hearing loss in humans
- Authors
- Vona, B., Mazaheri, N., Lin, S.J., Dunbar, L.A., Maroofian, R., Azaiez, H., Booth, K.T., Vitry, S., Rad, A., Rüschendorf, F., Varshney, P., Fowler, B., Beetz, C., Alagramam, K.N., Murphy, D., Shariati, G., Sedaghat, A., Houlden, H., Petree, C., VijayKumar, S., Smith, R.J.H., Haaf, T., El-Amraoui, A., Bowl, M.R., Varshney, G.K., Galehdari, H.
- Source
- Full text @ Hum. Genet.
|
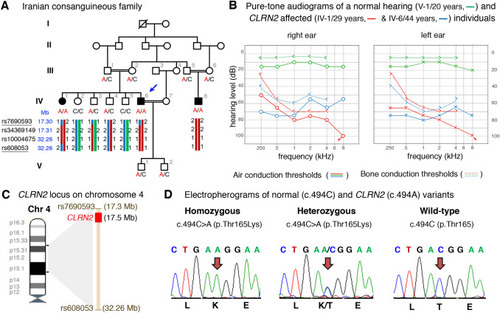
Pedigree, audiological data, genetic data, and locus mapping. |
|
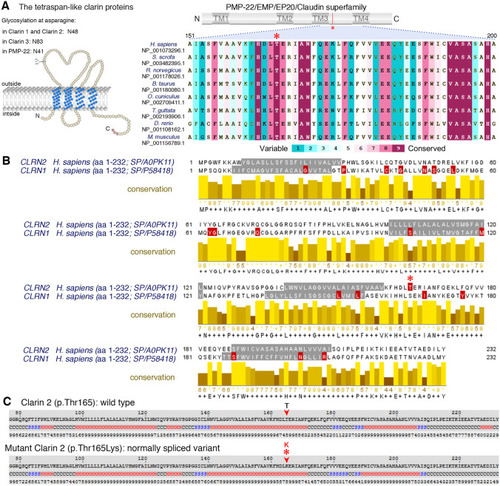
Conservation of the p.Thr165 residue, and clarin 1/clarin 2 alignment. |
|
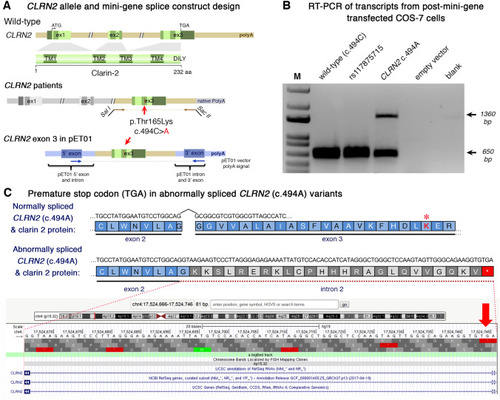
Analysis of the CLRN2 c.494C > A variant on splicing. a Schematic illustration of the mini-gene splice construct design. Genomic representation of CLRN2, including the position of the missense variant c.494C > A (arrow) on exon 3 with 3′ UTR (green), and the 5′ UTR, as well as exons 1 and 2 (grey) (upper panel). Regions captured by mini-gene PCR primers are represented in brown. Schematic illustration of the mini-gene splice construct including exon 3 and its flanking sequence (green) cloned into multiple cloning sites (SalI and SacII sites) of pET01 backbone vector (lower panel). Blue boxes represent native exons of the pET01 vector. b RT-PCR of transcripts from post-mini-gene transfected COS-7 cells. Amplicons derived from the transcripts of WT (CLRN2), a benign CLRN2 polymorphism (rs117875715, chr4(GRCh37):g.17,528,480G > A), the CLRN2 c.494C > A variant and a negative control, were visualized on a 1.5% agarose gel. The SNP, rs117875715, was used to test and validate the designed WT and mutant mini-gene assay. The ~ 650 bp amplicon was associated with the WT and validation control rs117875715. The amplicon derived from the CLRN2 c.494C > A transcripts showed two bands: a 650 bp band and a larger ~ 1360 bp band, indicating retention of intron separating the donor site of the 5′ exon and the acceptor site of CLRN2 exon 3. c Retention of intron in CLRN2 c.494C > A mini-gene results in a stop codon (TGA) after CLRN2 exon 2 |
|
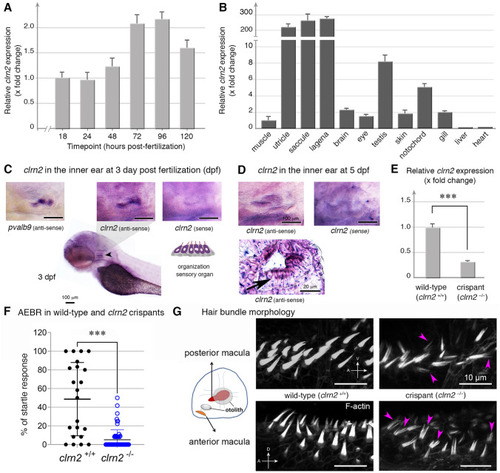
Clarin 2 is required for the inner ear function in zebrafish. EXPRESSION / LABELING:
|
|
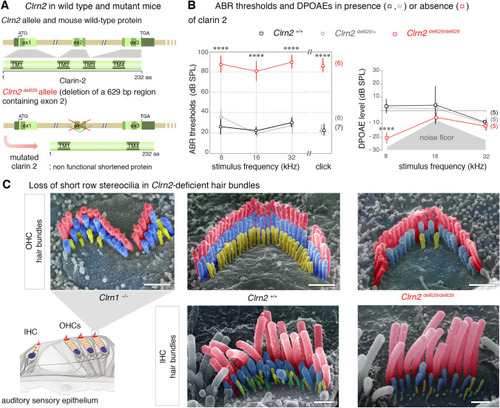
Clarin 2 is required for hearing function in mouse. |