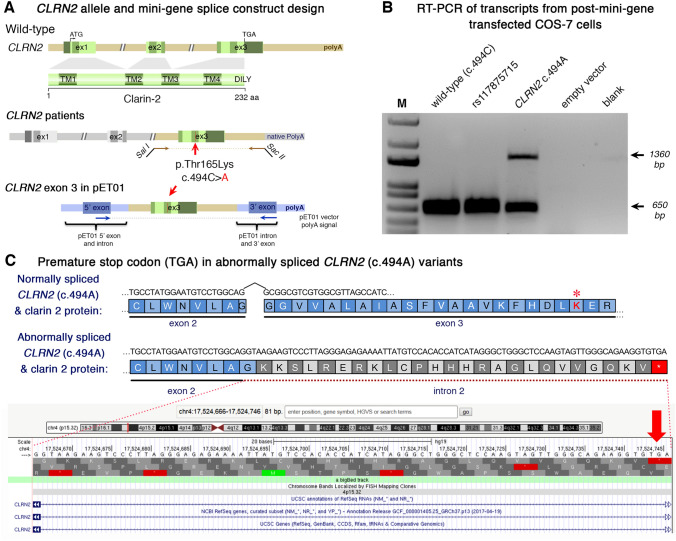
Fig. 3 Analysis of the CLRN2 c.494C > A variant on splicing. a Schematic illustration of the mini-gene splice construct design. Genomic representation of CLRN2, including the position of the missense variant c.494C > A (arrow) on exon 3 with 3′ UTR (green), and the 5′ UTR, as well as exons 1 and 2 (grey) (upper panel). Regions captured by mini-gene PCR primers are represented in brown. Schematic illustration of the mini-gene splice construct including exon 3 and its flanking sequence (green) cloned into multiple cloning sites (SalI and SacII sites) of pET01 backbone vector (lower panel). Blue boxes represent native exons of the pET01 vector. b RT-PCR of transcripts from post-mini-gene transfected COS-7 cells. Amplicons derived from the transcripts of WT (CLRN2), a benign CLRN2 polymorphism (rs117875715, chr4(GRCh37):g.17,528,480G > A), the CLRN2 c.494C > A variant and a negative control, were visualized on a 1.5% agarose gel. The SNP, rs117875715, was used to test and validate the designed WT and mutant mini-gene assay. The ~ 650 bp amplicon was associated with the WT and validation control rs117875715. The amplicon derived from the CLRN2 c.494C > A transcripts showed two bands: a 650 bp band and a larger ~ 1360 bp band, indicating retention of intron separating the donor site of the 5′ exon and the acceptor site of CLRN2 exon 3. c Retention of intron in CLRN2 c.494C > A mini-gene results in a stop codon (TGA) after CLRN2 exon 2
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Hum. Genet.