- Title
-
Zebrafish Model for Spondylo-Megaepiphyseal-Metaphyseal Dysplasia Reveals Post-Embryonic Roles of Nkx3.2 in the Skeleton
- Authors
- Smeeton, J., Natarajan, N., Naveen Kumar, A., Miyashita, T., Baddam, P., Fabian, P., Graf, D., Crump, J.G.
- Source
- Full text @ Development
|
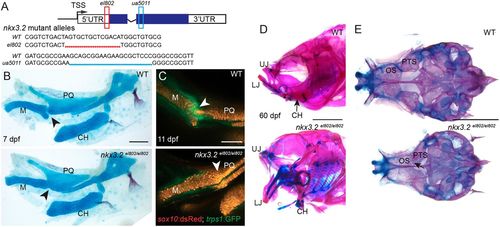
nkx3.2 mutant zebrafish are adult viable and develop craniofacial skeletal abnormalities. (A) Schematic of nkx3.2 mutant alleles: el802 – 14 bp deleted sequence including the ‘A’ of the ATG at the nkx3.2 translation start codon; ua5011 – 20 bp deleted sequence at the start of the homeobox DNA-binding domain. Coding regions of exons shown in blue and non-coding regions in white. (B) Lateral views of pharyngeal skeletal preparations stained with Alcian Blue (cartilage) and Alizarin Red (bone) show fusions of the jaw joint (arrowheads) in nkx3.2 mutants at 7 dpf. (C) Live imaging of transgenic juvenile zebrafish demonstrates loss of sox10:DsRed+/trps1:GFP+ jaw joint chondrocytes (arrowheads) in nkx3.2 mutants at 11 dpf. (D,E) Lateral views of the facial skeleton (D) and ventral views of the neurocranium (E) in adult nkx3.2 mutants at 60 dpf show craniofacial skeletal abnormalities including gaping jaw, fused jaw joint and displaced ceratohyal, and fusion of neurocranial orbitosphenoid and pterosphenoid bones (arrow in E). CH, ceratohyal; LJ, lower jaw; M, Meckel's cartilage; OS, orbitosphenoid; PQ, palatoquadrate; PTS, pterosphenoid; UJ, upper jaw; WT, wild type. Scale bars: 100 µm (B,C); 2 mm (D,E). EXPRESSION / LABELING:
PHENOTYPE:
|
|
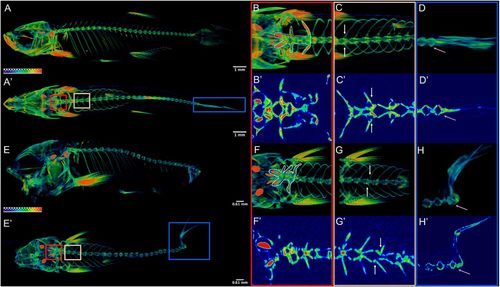
nkx3.2 mutant zebrafish develop axial skeletal abnormalities. (A,E) µCT imaging of 60 dpf wild types (A,A′) and nkx3.2ua5011/ua5011 mutants (E,E′) shown with gradient 3D rendering in sagittal (A,E) and dorsal (A′,E′) views. (B-D,F-H) Magnifications of three spinal regions: anterior vertebrae (red box, inclusive of the occiput and Weberian apparatus: B,B′,F,F′), precaudal vertebrae with rib articulations (tan box, white arrows indicate corresponding left and right ribs: C,C′,G,G′), and caudal fin vertebrae (blue box, pink arrow indicates caudal-most vertebra: D,D′,H,H′) shown in 3D (B,C,D,F,G,H) and 2D (B′,C′,D′,F′,G′,H′) slice. Scale bars: 1 mm (A,A′); 0.61 mm (E,E′). PHENOTYPE:
|
|
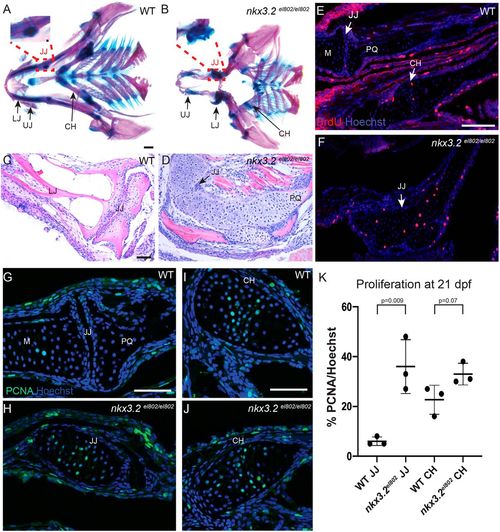
Cartilage overgrowth at the jaw joint in nkx3.2 mutant zebrafish. (A,B) Ventral views of dissected pharyngeal skeletons stained with Alcian Blue (cartilage) and Alizarin Red (bone) at 60 dpf. Insets with further dissections of the boxed regions in A and B demonstrate the wild-type retroarticular cartilage in A and the cartilage overgrowth at the site of the fused jaw joint in nkx3.2 mutants in B. (C,D) Frontal H&E-stained sections through 60 dpf jaw joints show fusion of the jaw joint and cartilage overgrowth in nkx3.2 mutants. (E-J) In wild types at 21 dpf (E,G,I), PCNA+ cells are present largely on the lower jaw side of the jaw joint, and PCNA+ and BrdU+ chondrocytes are present in the proliferative zone of the ceratohyal. In nkx3.2el802 mutants (F,H,J), proliferating chondrocytes marked by BrdU (E,F) and PCNA (G-J) are present across the fused jaw joint region. (K) Quantification of PCNA+ cells per total nuclei stained with Hoechst. Individual animals are plotted (dots) with mean±s.d. CH, ceratohyal; JJ, jaw joint; LJ, lower jaw; M, Meckel's cartilage; PQ, palatoquadrate; UJ, upper jaw; WT, wild type. Scale bars: 500 µm (A,B); 50 µm (C-J). PHENOTYPE:
|
|
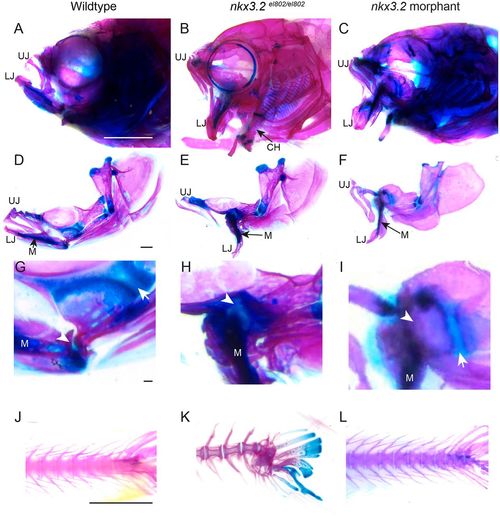
Adult nkx3.2 morphants lack the cartilage overgrowth and spine defects of nkx3.2 mutants. (A-I) Lateral views of whole-mount (A-C) and dissected (D-F) facial skeletons at 60 dpf. Alcian Blue (cartilage) and Alizarin Red (bone) staining show similar gaping jaws in mutant and morphant animals. High magnification views of the jaw joint (G-I) show ectopic cartilage across the fused jaw joint domain (arrowheads) in mutants (H) but not morphants (I). Arrows indicate distinct palatoquadrate growth plates present in wild types and morphants but not in mutants. (J-L) Lateral views of the caudal spine stained with Alcian Blue and Alizarin Red show spinal curvature in mutants but not in morphants. CH, ceratohyal; LJ, lower jaw; M, Meckel's cartilage; UJ, upper jaw. Scale bars: 2 mm (A-C; J-L); 500 µm (D-F); 50 µm (G-I). PHENOTYPE:
|
|
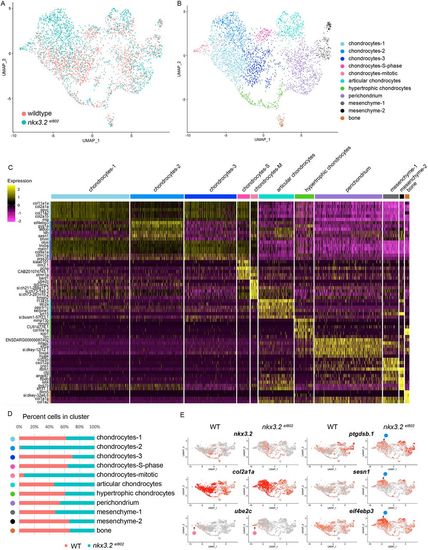
Single-cell analysis reveals altered facial chondrocyte subtypes in nkx3.2 mutants. (A,B) UMAP projections of the aggregate datasets colored according to dataset (A) or cluster identity (B). (C) Heatmap of the top 4-6 enriched genes per cluster. Yellow indicates high expression and magenta minimal. (D) Percent contribution within each cluster by control (WT) and mutant cells. (E) UMAP projections of gene expression split by genotype. Red indicates high expression and grey minimal. Pink circles indicate position of the mitotic chondrocytes cluster and blue circles indicate the chondrocytes-2 cluster. |
|
Increased expression of ptgdsb.1 and sesn1 across the fused jaw joint of nkx3.2 mutant zebrafish. (A,B) Colorimetric in situ hybridization demonstrates nkx3.2 expression in the sub-articular zone of the jaw joint and across the fused jaw joint in nkx3.2el802 mutants at 21 dpf. (C-F) Colorimetric in situ hybridization shows expansion of ptgdsb.1 but not col10a1a across the fused jaw joint region of nkx3.2 mutants. (G-J′) RNAScope in situ hybridization shows ectopic expression of ptgdsb.1 (G-H′) and sesn1 (I-J′) across the fused jaw joint region of mutants (identified by nkx3.2 expression in red). DAPI labels nuclei in blue. Dashed lines in H,H′ indicate fused jaw joint region. (K-P) RNAscope in situ hybridizations show expression of col10a1a, ptgdsb.1, nkx3.2 and sesn1 in the growth plates of the ceratohyal. (Q) Quantification of fluorescence intensity at the jaw joint demonstrates upregulation of ptgdsb.1 and sesn1 in nkx3.2 mutants. Individual animals are plotted (dots) with mean±s.d. (R,S) Model of chondrocyte zones at the wild-type and fused mutant jaw joint (R) and within the bidirectional ceratohyal growth plate (S). AC, articular surface; Hyp, hypertrophic; JJ, jaw joint; LH, late hypertrophic; M, Meckel's cartilage; PH, pre-hypertrophic; PQ, palatoquadrate; PZ, proliferative zone; SA, sub-articular zone; WT, wild type. Scale bars: 50 µm. EXPRESSION / LABELING:
PHENOTYPE:
|