- Title
-
Phosphorylation of seryl-tRNA synthetase by ATM/ATR is essential for hypoxia-induced angiogenesis
- Authors
- Shi, Y., Liu, Z., Zhang, Q., Vallee, I., Mo, Z., Kishi, S., Yang, X.L.
- Source
- Full text @ PLoS Biol.
|
( |
|
( EXPRESSION / LABELING:
PHENOTYPE:
|
|
( |
|
( |
|
( |
|
( |
|
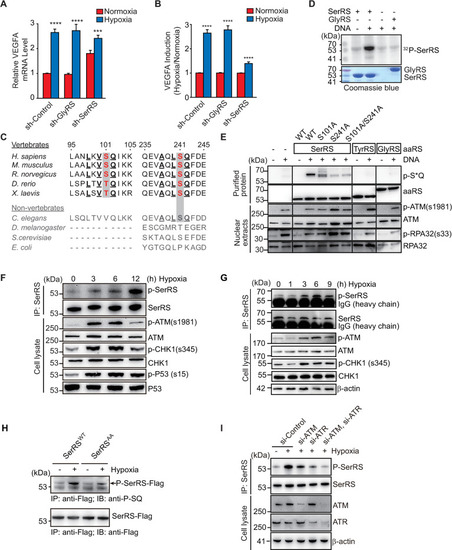
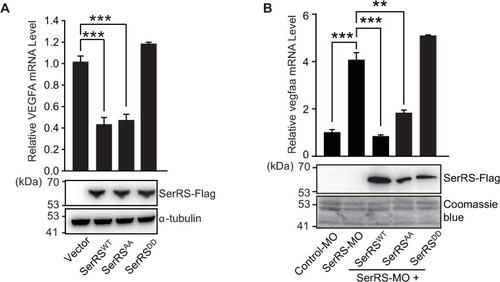
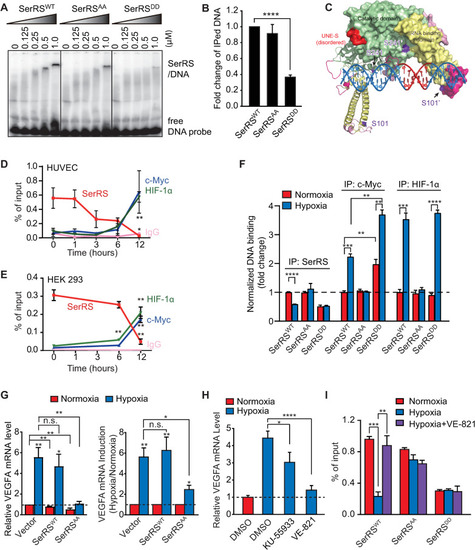
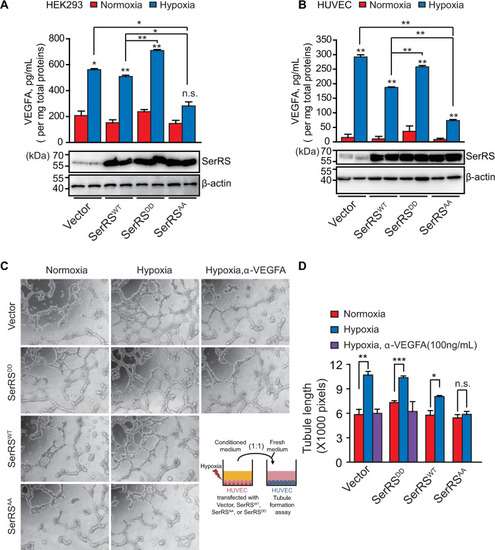
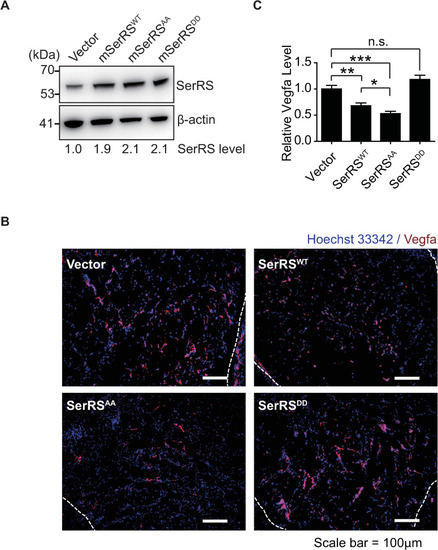
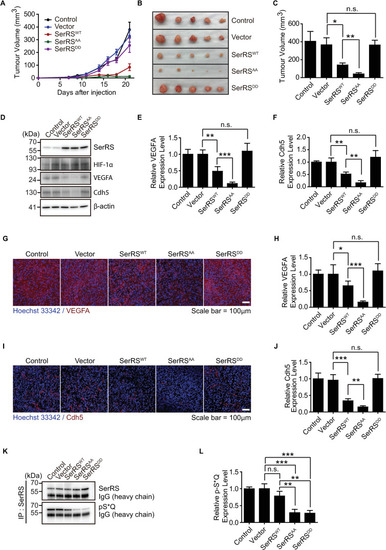
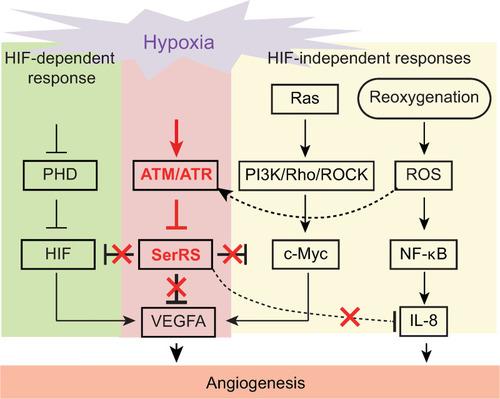
Multiple pathways are activated under hypoxia to induce angiogenesis. SerRS has the capacity to inhibit these pathways regardless of whether they are dependent or independent of HIF. This capacity, however, is silenced by ATM/ATR, which is also activated by hypoxia, highlighting the central role of SerRS in regulating angiogenesis. ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia mutated and RAD3-related; HIF; hypoxia-inducible factor; SerRS, seryl-tRNA synthetase. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|