- Title
-
Rad21 Haploinsufficiency Prevents ALT-Associated Phenotypes in Zebrafish Brain Tumors
- Authors
- Idilli, A.I., Pazzi, C., Dal Pozzolo, F., Roccuzzo, M., Mione, M.C.
- Source
- Full text @ Genes (Basel)
|
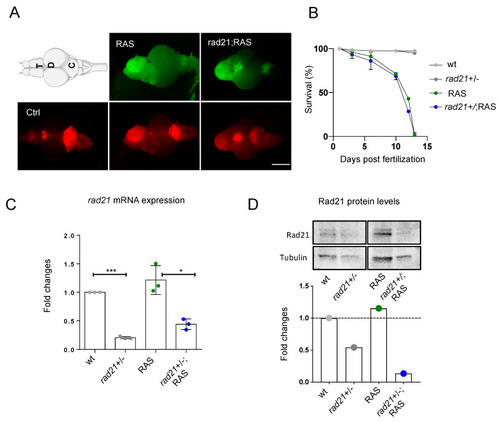
Rad21 heterozygous fish develop brain tumors with the same frequency and location than wt. ( |
|
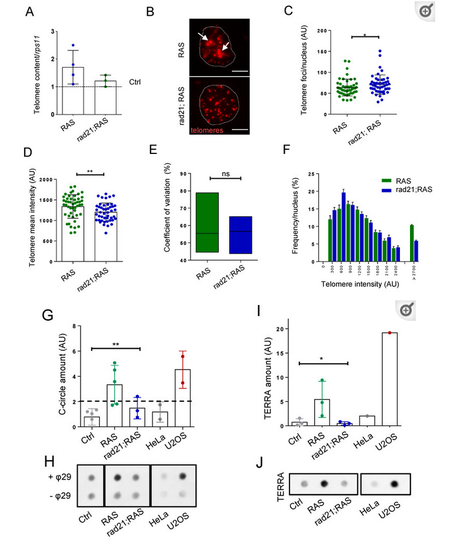
Rad21 depletion prevents ALT, normalize telomere length and reduces TERRA levels in brain tumors. ( PHENOTYPE:
|
|
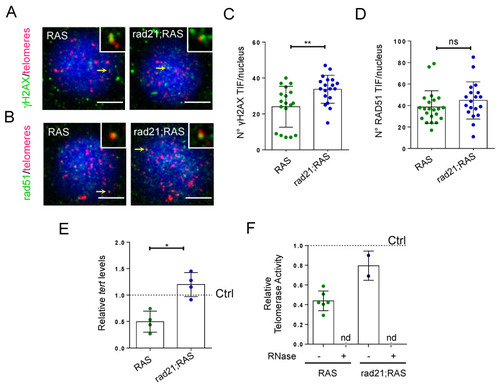
Rad21 depletion increases |