- Title
-
Functional Characterization of the N-Acetylmuramyl-l-Alanine Amidase, Ami1, from Mycobacterium abscessus
- Authors
- Küssau, T., Van Wyk, N., Johansen, M.D., Alsarraf, H.M.A.B., Neyret, A., Hamela, C., Sørensen, K.K., Thygesen, M.B., Beauvineau, C., Kremer, L., Blaise, M.
- Source
- Full text @ Cells
|
Generation and phenotypic analyses of Δ |
|
Assessment of Δ PHENOTYPE:
|
|
Biochemical characterization of Ami1Mab. ( |
|
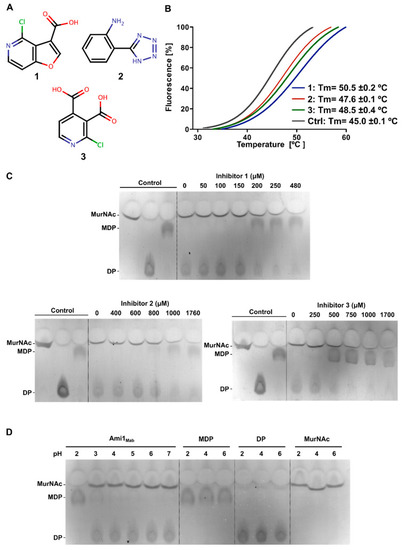
Characterization of Ami1Mab inhibitors. ( |
|
Structural comparisons of the Ami1Mab, Ami1Mtb, and Ami1Msm. ( |
|
In silico docking of the three inhibitors. The figure depicts the best docking poses of the three compounds. All three inhibitors appear able to bind in the Ami1Mab active site with similar binding energies. Inhibitors are displayed in stick representation. Nitrogen atoms are in blue, oxygen in red, hydrogen in white and carbon atoms in yellow for the ligand and grey for the protein. |