- Title
-
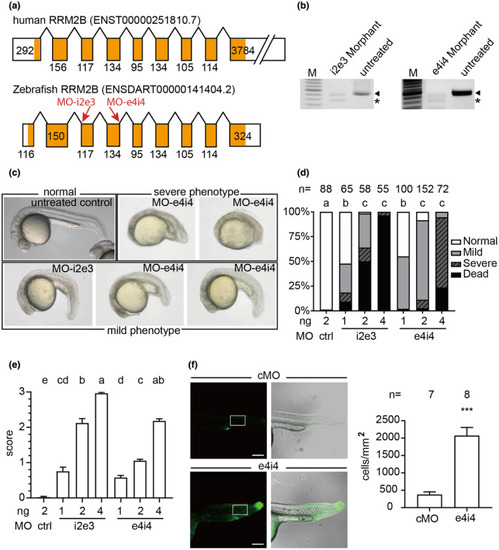
The single nucleotide variant at c.662A>G in human RRM2B is a loss-of-function mutation
- Authors
- Tseng, Y.T., Li, S.W., HuangFu, W.C., Yen, Y., Liu, I.H.
- Source
- Full text @ Mol Genet Genomic Med

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Phenotypic characterization of PHENOTYPE:
|