- Title
-
Systems genetics analysis identifies calcium-signaling defects as novel cause of congenital heart disease
- Authors
- Izarzugaza, J.M.G., Ellesøe, S.G., Doganli, C., Ehlers, N.S., Dalgaard, M.D., Audain, E., Dombrowsky, G., Banasik, K., Sifrim, A., Wilsdon, A., Thienpont, B., Breckpot, J., Gewillig, M., Competence Network for Congenital Heart Defects, Germany, Brook, J.D., Hitz, M.P., Larsen, L.A., Brunak, S.
- Source
- Full text @ Genome Med.
|
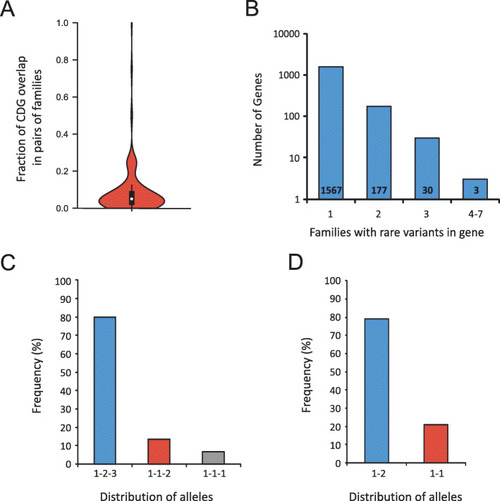
Distribution of candidate disease genes and variants across families. |
|
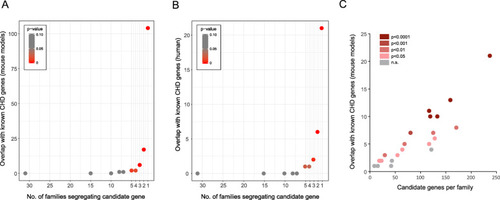
Enrichment of CHD genes in candidate disease genes. Overlap between CDGs and known CHD genes from mouse models ( |
|
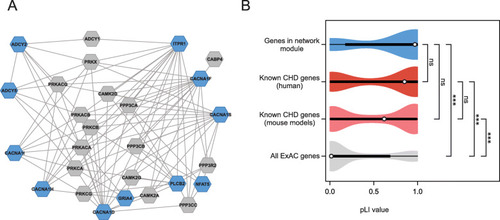
Identification of a calcium-signaling network affected by rare mutations identified in CHD families. |
|
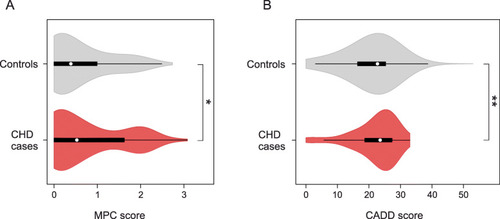
Distribution of MPC and CADD scores of rare variants in 714 CHD cases and 4922 controls. Protein altering and truncating variants (PAV and PTV) with MAF < 0.001 identified in the genes |
|
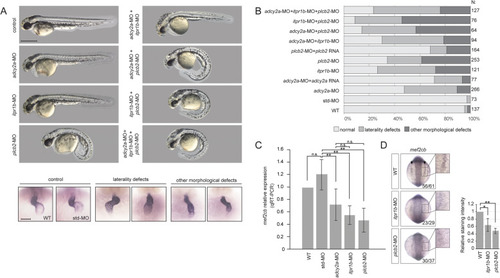
Functional validation of candidate genes in zebrafish. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|