- Title
-
Bi-FoRe: an efficient bidirectional knockin strategy to generate pairwise conditional alleles with fluorescent indicators
- Authors
- Han, B., Zhang, Y., Bi, X., Zhou, Y., Krueger, C.J., Hu, X., Zhu, Z., Tong, X., Zhang, B.
- Source
- Full text @ Protein Cell
|
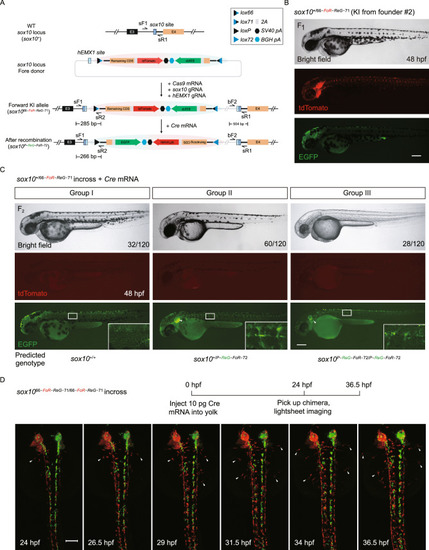
Generation and evaluation of fluorescent reporter-tagged conditional knockout alleles at the zebrafish |
|
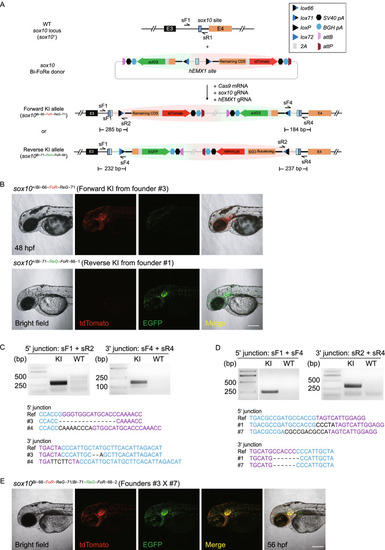
Generation of positive and negative conditional allele pairs at the |
|
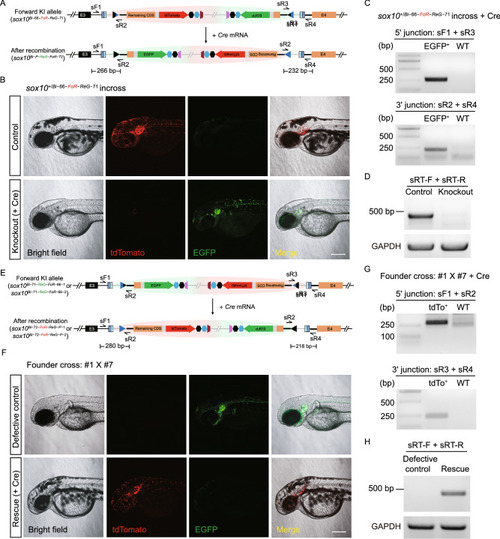
Evaluation of conditional manipulation of paired positive and negative conditional Bi-FoRe KI alleles at the |
|
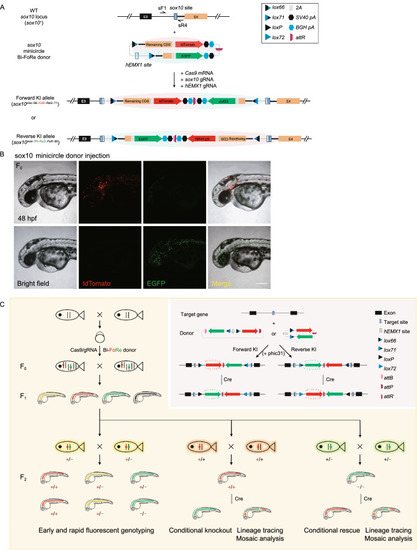
Generation of conditional |