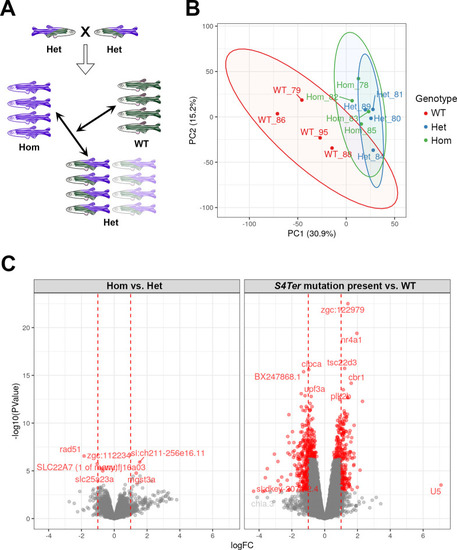
- Title
-
Transcriptome analysis indicates dominant effects on ribosome and mitochondrial function of a premature termination codon mutation in the zebrafish gene psen2
- Authors
- Jiang, H., Pederson, S.M., Newman, M., Dong, Y., Barthelson, K., Lardelli, M.
- Source
- Full text @ PLoS One
|
PHENOTYPE:
|
|
|
|
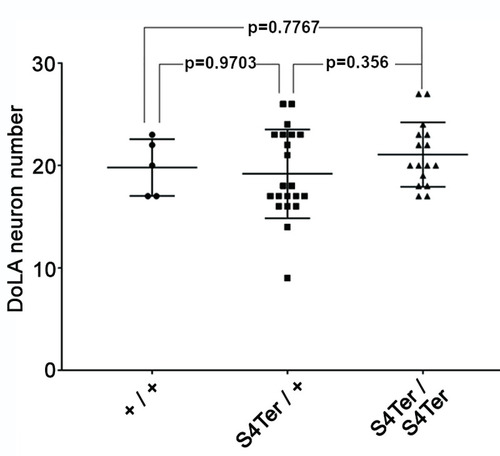
42 embryos at 24 hpf from a pair-mating of a PHENOTYPE:
|
|
|
|
|
|
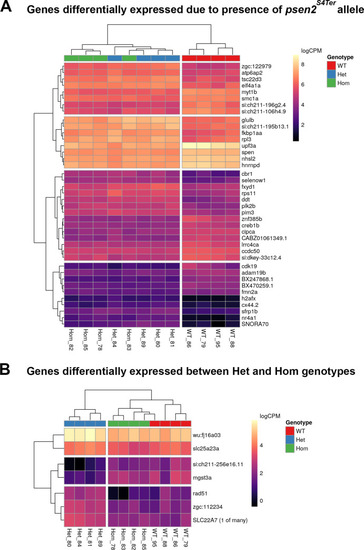
Plotted values are logCPM based on CQN-normalized counts. |
|
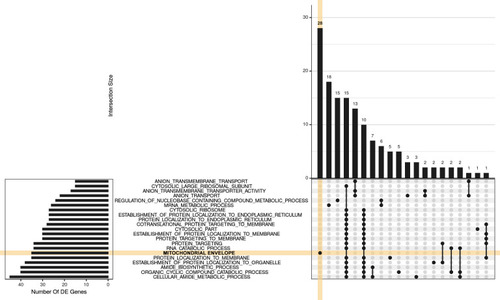
For this visualization, GO terms were restricted to those with 15 or more DE genes, where this represented more than 5% of the gene set, along with an FDR < 0.02 and more than 3 steps back to the ontology root. The 20 largest GO terms satisfying these criteria are shown with the plot being truncated at the right hand side for simplicity. A group of 28 genes is uniquely attributed to the GO |