- Title
-
Swimming motility of a gut bacterial symbiont promotes resistance to intestinal expulsion and enhances inflammation
- Authors
- Wiles, T.J., Schlomann, B.H., Wall, E.S., Betancourt, R., Parthasarathy, R., Guillemin, K.
- Source
- Full text @ PLoS Biol.
|
(A) Abundances of wt |
|
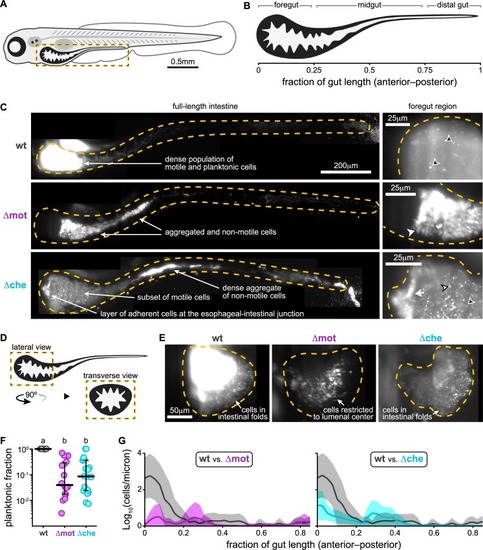
(A) Cartoon of a 6-day-old zebrafish. Dashed box marks intestinal region imaged by LSFM. (B) Anatomical regions of the larval zebrafish intestine. (C) Maximum intensity projections acquired by LSFM showing the spatial organization of wt |
|
(A) Image-based quantification of abundances over time for wt |
|
(A) Schematic of CRISPRi-based motility LOF switch. Lower left table summarizes switch activity and bacterial behaviors +/− aTc. Bent arrows denote promoters; “T” denotes transcriptional terminators. Solid lines represent constitutive interactions; dashed lines represent induced interactions. (B) Experimental timelines used to investigate in situ inactivation of swimming motility. (C) A maximum intensity projection acquired by LSFM of an animal colonized by |
|
(A) Schematic of the motility and chemotaxis GOF switches. Table summarizes switch activity and bacterial behaviors +/− aTc. (B) ΔmotGOF or ΔcheGOF abundances 24 hpi +/− aTc. ΔmotGOF and ΔcheGOF were preinduced overnight in liquid culture prior to inoculation; aTc was maintained in the water for continuous switch activation. Abundances of wild-type |
|
(A) Maximum intensity projections acquired by LSFM of the foregut region of |
|
(A) Maximum intensity projections acquired by LSFM of the foregut region of separate |