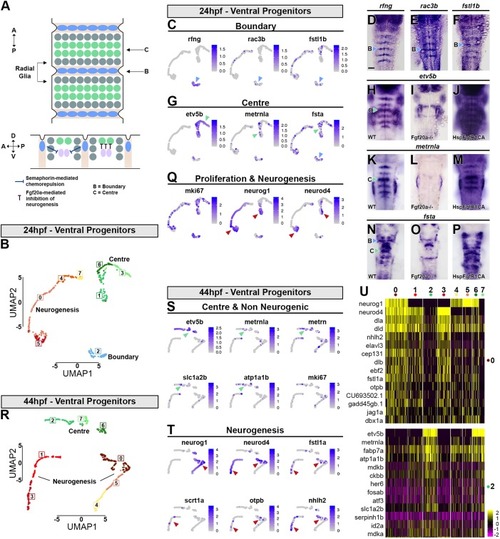
Transcriptional signature of boundary cells and segment centre progenitors. (A) Schematic drawing representing anterior-posterior organisation within hindbrain segments. Boundary cells are in blue, neurogenic progenitors in grey and segment centre cells in green. Below is a side view showing the role of boundary cells in maintaining Fgf20a neurons (pink) at the centre of each segment, mediated by semaphorins. Fgf20 signalling maintains undifferentiated progenitors. (B) Supervised clustering of 24 hpf ventral progenitors. Eight clusters are identified: C7, C4 and C0 are progenitors; C5 is the neurogenic domain; C2 are boundary cells; and C1, C6 and C3 are segment centre progenitors. (C,G,Q) UMAP plots showing the expression distribution of boundary (C), segment centre (G) and proliferation and neurogenic genes (Q). Arrowheads indicate relevant domain of expression; colour refers to cluster of origin. (D-F,H,K,N) Whole-mount in situ hybridisation of boundary (D-F) and segment centre genes (H,K,N). (I,J,L,M,O,P) Segment centre-specific gene expression is dependent on Fgf20 signalling, as fgf20a−/− embryos have loss of etv5b (I), metrnla (L) and fsta (O) expression, whereas constitutive activation of FgfR1 induces their ectopic expression (J,M,P). (R) Supervised clustering of 44 hpf ventral progenitors. Eight clusters are identified: C4 and C5 are progenitors; C0, C1 and C3 are neurogenic domains; C2, C7 and C6 are segment centre progenitors. (S,T) UMAP plots showing the expression distribution of segment centre and non-neurogenic genes (S) and neurogenic genes (T). Arrowheads indicate relevant domain of expression; colour refers to cluster of origin. (U) Heatmap of the top 15 genes enriched in each cluster.
|