- Title
-
Elongator Subunit 3 (Elp3) Is Required for Zebrafish Trunk Development
- Authors
- Rojas-Benítez, D., L Allende, M.
- Source
- Full text @ Int. J. Mol. Sci.
|
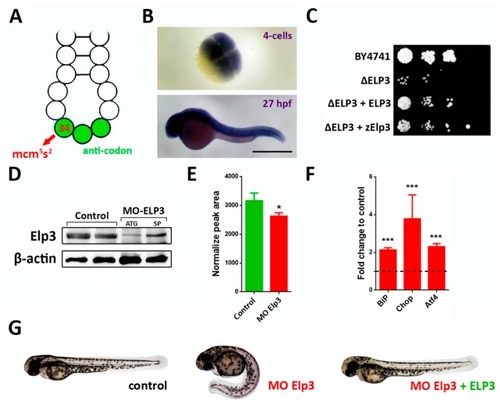
Elp3 morphants present a ventrally-curved tail. ( |
|
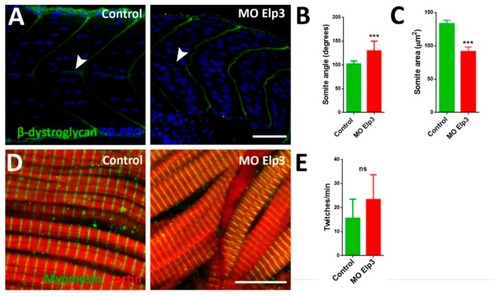
Aberrant somite shape and horizontal myoseptum in Elp3 morphants. To show somite boundary we detected β-dystroglycan in ( PHENOTYPE:
|
|
Abnormal muscle fiber morphology in morphants. Muscle fibers of control ( |
|
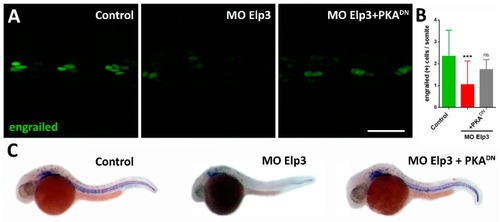
Shh activity is diminished in ELP morphants. ( |
|
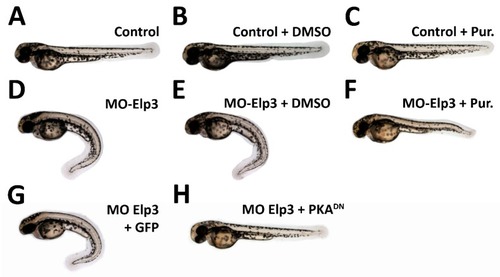
Morphant phenotype is rescued by sonic hedgehog pathway activation. Images of 2dpf larvae of ( PHENOTYPE:
|