- Title
-
The Autocrine FGF/FGFR System in both Skin and Uveal Melanoma: FGF Trapping as a Possible Therapeutic Approach
- Authors
- Rezzola, S., Ronca, R., Loda, A., Nawaz, M.I., Tobia, C., Paganini, G., Maccarinelli, F., Giacomini, A., Semeraro, F., Mor, M., Presta, M.
- Source
- Full text @ Cancers
|
Fibroblast growth factor receptor (FGFR) and fibroblast growth factor (FGF) overexpression in human primary uveal melanoma (UM). Analysis of The Cancer Genome Atlas (TCGA) dataset was performed on a cohort of 80 UM patients. ( |
|
Correlation between FGF/FGFR expression and chromosome 3 / |
|
Effect of long-pentraxin 3 (PTX3) overexpression on B16-LS9 cells. ( |
|
Effect of the pan FGF-trap NSC12 on B16-LS9 cells. ( |
|
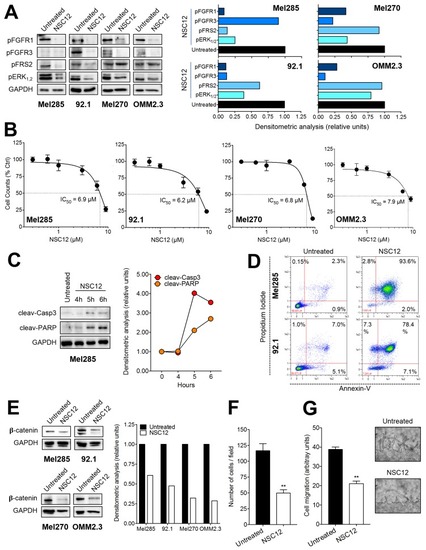
Effect of the pan FGF-trap NSC12 on human UM cells. ( |