- Title
-
AMPK promotes induction of the tumor suppressor FLCN through activation of TFEB independently of mTOR
- Authors
- Collodet, C., Foretz, M., Deak, M., Bultot, L., Metairon, S., Viollet, B., Lefebvre, G., Raymond, F., Parisi, A., Civiletto, G., Gut, P., Descombes, P., Sakamoto, K.
- Source
- Full text @ FASEB J.
|
AMPK-activation profiles in samples used for transcriptome analyses. |
|
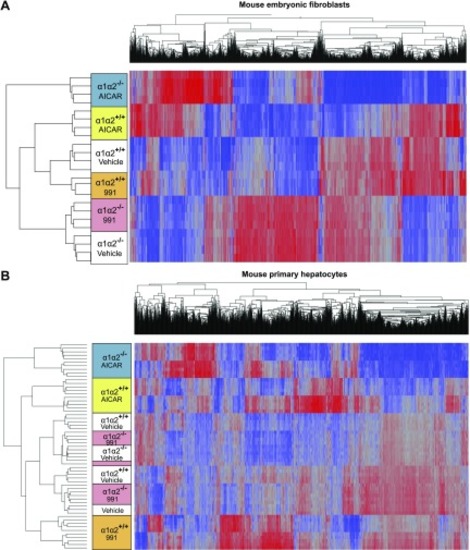
Hierarchical clustering of the transcriptome data. Overview of the hierarchical clustering analyses in MEFs ( |
|
Transcriptome data analysis of the AMPK-activation response following treatment with 991 and AICAR. Venn diagrams showing the transcriptome profiling specificity of 991 and AICAR in MEFs ( |
|
Identification of pathways and genes modulated by AMPK. |
|
Identification of |
|
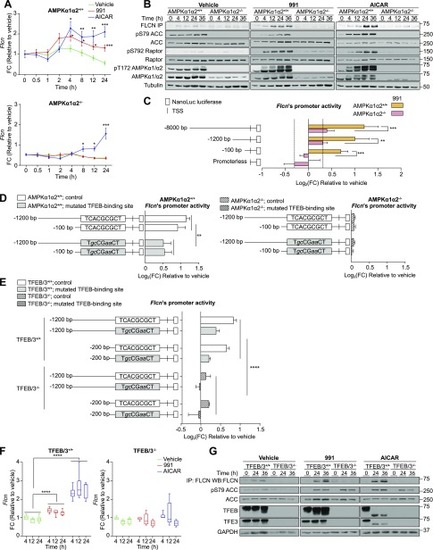
Pharmacological activation of AMPK leads to dephosphorylation and nuclear localization of TFEB. |
|
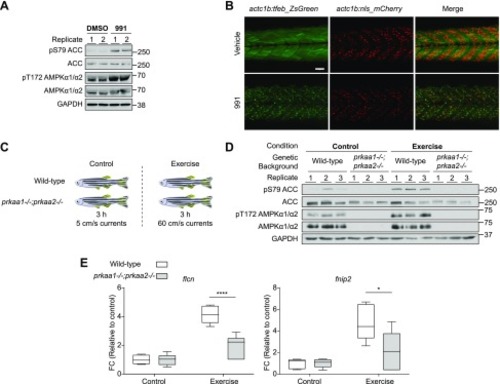
TFEB translocates to the nucleus upon AMPK stimulation independently of mTOR in mouse primary hepatocytes. |
|
Activation of AMPK leads to translocation of TFEB to the nucleus and increased expression of |