- Title
-
Specific inhibition of splicing factor activity by decoy RNA oligonucleotides
- Authors
- Denichenko, P., Mogilevsky, M., Cléry, A., Welte, T., Biran, J., Shimshon, O., Barnabas, G.D., Danan-Gotthold, M., Kumar, S., Yavin, E., Levanon, E.Y., Allain, F.H., Geiger, T., Levkowitz, G., Karni, R.
- Source
- Full text @ Nat. Commun.
|
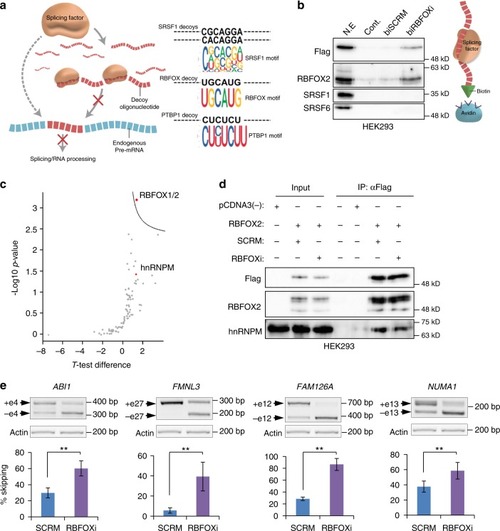
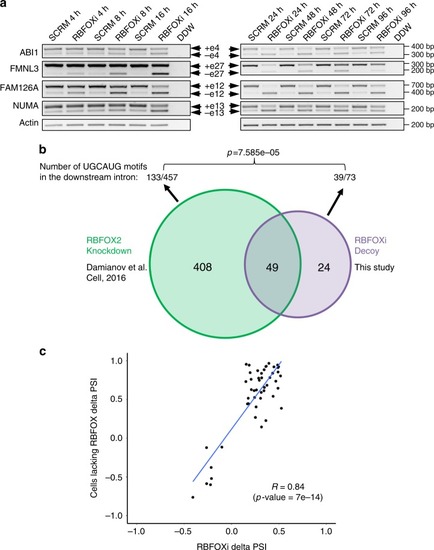
RBFOX1/2 decoy oligonucleotide binds RBFOX2 and affects its splicing targets. |
|
RNA-seq of RBFOXi transfected U87MG cells. |
|
RBFOX1 RRM interaction with RNAs containing consecutive binding sites using switchSENSE. |
|
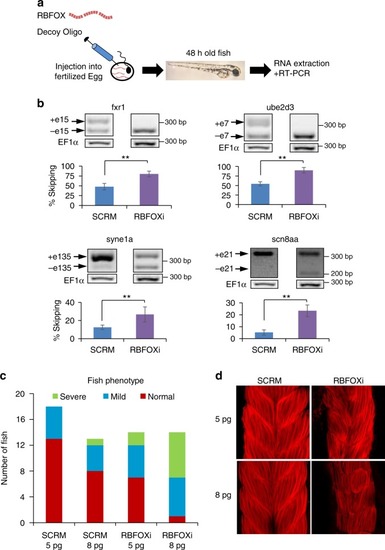
RBFOXi affects alternative splicing and muscle development in zebrafish. |
|
PTBP1 decoy oligonucleotides inhibit its splicing and biological activities in breast cancer cells. |
|
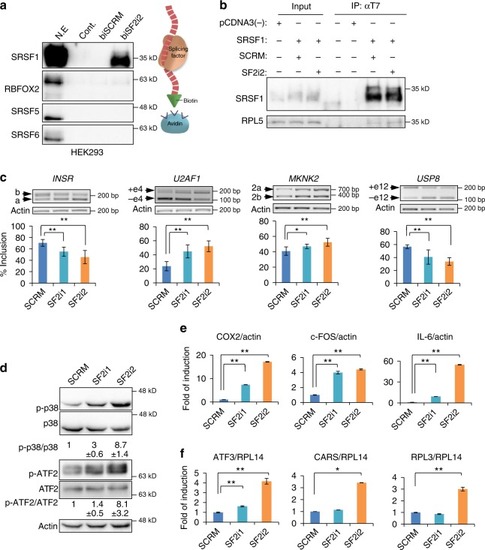
SRSF1 decoy oligonucleotides inhibit splicing and biological functions in glioblastoma cells. |
|
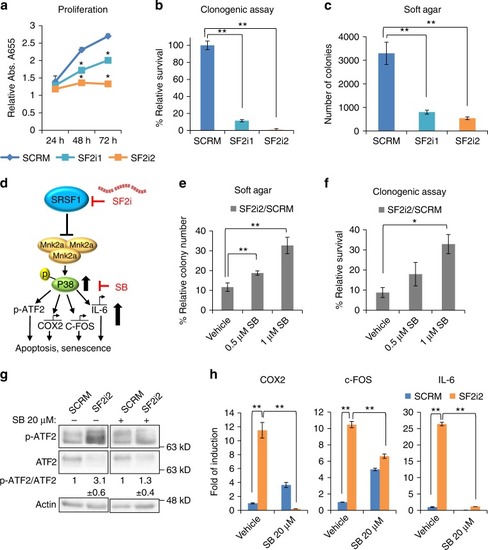
Inhibition of glioblastoma cells by SRSF1 decoys is partially reversed by SB203580. |
|
SRSF1 decoy oligonucleotides inhibit glioblastoma tumor growth in mice. |