- Title
-
Leveraging Zebrafish to Study Retinal Degenerations
- Authors
- Angueyra, J.M., Kindt, K.S.
- Source
- Full text @ Front Cell Dev Biol
|
Structure of the zebrafish eye and retina. |
|
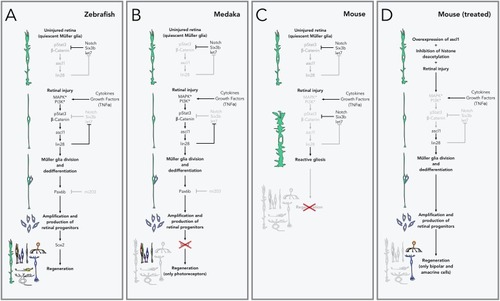
Pathways to retinal regeneration. |
|
Tools to study retinal circuits. |