- Title
-
ALPK2 Promotes Cardiogenesis in Zebrafish and Human Pluripotent Stem Cells
- Authors
- Hofsteen, P., Robitaille, A.M., Strash, N., Palpant, N., Moon, R.T., Pabon, L., Murry, C.E.
- Source
- Full text @ iScience
|
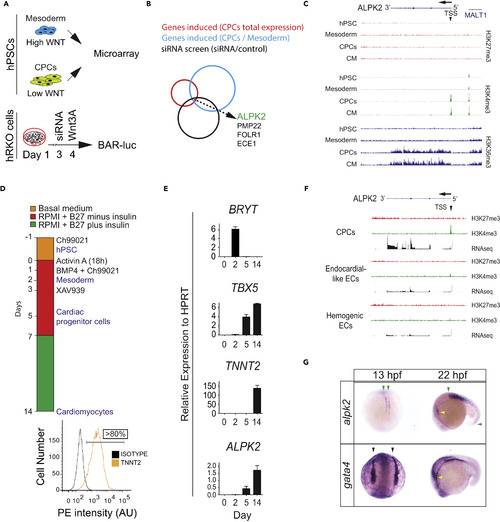
ALPK2 Identification and Expression Analyses (A) Schematic of combinatorial screening of RNA expression from hESC cardiomyocyte differentiation from human embryonic stem cells (hESCs) coupled with an siRNA screen using β-catenin-activated reporter (BAR)-transduced human RKO colon carcinoma cells (hRKO) stimulated with recombinant Wnt3A. (B) Venn diagram identifying Alpha Protein Kinase 2 (ALPK2). (C) Chromatin precipitation followed by deep sequencing (ChIP-Seq) for histone marks H3K4me3, H3K36me3, and H27K4me3 in hESC-derived cultures: hESC (day 0), mesoderm (day 2), cardiac progenitor cells (CPCs, day 5), and cardiomyocytes (day 14) (N = 2). (D) Protocol for high-density monolayer-directed differentiation of hESC-derived cardiomyocytes yielding a high percentage of cardiac troponin T (TNNT2)-positive cells by flow cytometry. (E) Quantitative RT-PCR analysis of markers of mesoderm (Brachyury T, BRYT), cardiac progenitor cells (T-box 5, TBX5), cardiomyocytes (TNNT2), and ALPK2 at days 0, 2, 5, 14 during cardiomyocyte differentiation. (F) RNA sequencing and ChIP-seq for H3K4me3 and H27K4me3 at the ALPK2 locus in CPCs, endocardial-like endothelial cells (EC), and hemogenic ECs (N = 2). (G) In situ hybridization of zebrafish alpk2 and gata4 at 13 hpf and 22 hr post fertilization (hpf, N = 22–36). Green arrowheads denote adaxial cells and primitive somites, black arrowheads mark the bilateral heart fields, and yellow arrowheads denote the primitive heart. Sample size N = 3–5 biological replicates, and data are displayed as mean ± SEM unless otherwise noted. See also Figure S1. EXPRESSION / LABELING:
|
|
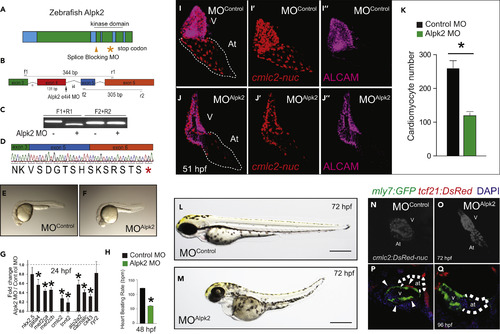
Alpk2 Is Essential for Zebrafish Cardiac Development (A) Zebrafish Alpk2 locus and targeted region by splice-blocking morpholinooligonucleotides (MO). * denotes stop codon. (B–D) RT-PCR and Sanger sequencing demonstrating splice-blocking MO caused alternative splicing resulting in a 131-base pair deletion and premature stop codon. * denotes stop codon. (E and F) Zebrafish injected with negative control MO (E) or Alpk2 MO (F) at 24 hr post fertilization (hpf). (G and H) Transcript analysis (G) and heart beating rate (H) of control and Alpk2 morphants at 24 and 48 hpf, respectively (N = 5, 15–22 pooled embryos per N). (I–O) Representative images of control and Alpk2 MO-injected hearts carrying a transgene for cmlc2:DsRed-nuc to quantify cardiomyocyte nuclei (K) at 51 hpf (N = 7–9). Red denotes cmlc2:DsRed-nuc and magenta is antibody staining for activated leukocyte cell adhesion molecule (ALCAM). Representative bright-field images of control MO (L) and Alpk2 MO (M) injected zebrafish at approximately 72 hpf. Representative images of control MO (N) and Alpk2 MO (O) injected zebrafish hearts carrying a transgene (cmlc2:DsRed-nuc) to denote cardiac morphology at approximately 72 hpf. (P and Q) Confocal images of control and Alpk2 morphant fish carrying myl7:GFP and tcf21:DsRed at 96 hpf. White arrowheads denote tcf21+ epicardial cells on the heart ventricle. N = 3; independent biological replicates with 45–76 pooled animals per N unless otherwise noted. Data are mean ± SEM; * denotes p ≤ 0.05. |
|
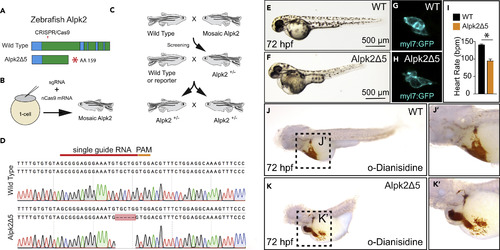
CRISPR/Cas9 Knockout of Zebrafish Alpk2 Inhibits Cardiac Development (A) Schematic of the zebrafish Alpk2 locus and CRISPR/Cas9 targeted region. * denote stop codon. (B and C) Injection and breeding paradigm to generate Alpk2 null homozygosity. (D–H) (D) Sanger sequencing of homozygous mutation showing 5-bp deletion. Representative bright-field (E, F; 72 hpf) and fluorescent (G, H; 120 hpf) images of wild-type (E, G) and Alpk2 null zebrafish (F, H) carrying a transgene for myl7:GFP (G, H). (I) Quantification of cardiac beating rate comparing Alpk2 null zebrafish to wild-type siblings at 72 hpf. * denotes p≤0.05. (J and K) Representative bright-field images of hemoglobin-peroxidase staining with o-Dianisidine in 72 hpf WT and homozygous Alpk2Δ5 embryos. Data are representative of N ≥ 3 independent breeding experiments consisting of n∼30–150 embryos per spawn. hpf, hours post fertilization. PHENOTYPE:
|
|
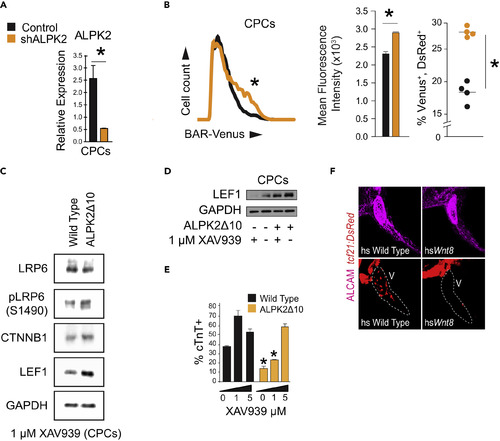
ALPK2 Inhibits WNT/β-Catenin Signaling to Promote Cardiogenesis (A) Transcripts abundance of ALPK2 in control and ALPK2 shRNA-transfected cardiac progenitor cells (CPCs) derived from human embryonic stem cells (hESCs). (B) Flow cytometric plot and graphs quantifying venus expression driven by β-catenin activated reporter (BAR) in live CPCs following control or ALPK2 shRNA transfection as hESCs. (C) Western blot protein expression analysis of WNT/β-catenin signaling modulators in wild-type and CRISPR/Cas9-mediated ALPK2 mutant hESC-derived CPCs (ALPK2Δ10) treated with a small molecule direct tankyrase inhibitor XAV939 (1 μM). (D) Western blotting for the WNT/β-catenin signaling target LEF1 in wild-type and ALPK2Δ10 cells with or without the addition of XAV939 (0 or 1 μM). (E) TNNT2 flow cytometry quantification of day 14 cardiomyocytes derived from wild-type and ALPK2Δ10 hESCs treated with XAV939 (0, 1, 5 μM). (F) Representative images of heat shocked (hs) wild-type and hsWnt8b transgenic zebrafish carrying a transgene for the epicardium (tcf21:DsRed) at 96 hpf. Fish were counterstained with an activated leukocyte cell adhesion molecule (ALCAM; magenta). N = 3–5 biological replicates, and data are displayed as mean ± SEM. * = p ≤ 0.05. See also Figure S4. PHENOTYPE:
|