- Title
-
Rare copy number variants analysis identifies novel candidate genes in heterotaxy syndrome patients with congenital heart defects
- Authors
- Liu, C., Cao, R., Xu, Y., Li, T., Li, F., Chen, S., Xu, R., Sun, K.
- Source
- Full text @ Genome Med.
|
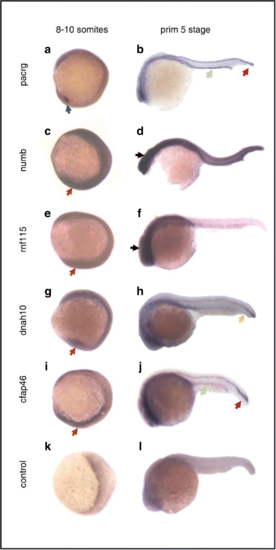
Whole mount in situ hybridization analysis of candidate genes at two stages: 8–10 somites and primordium 5 stage. a, c, e, g, i, k Results of in situ hybridization of candidate genes and standard control at 13–15 hpf (8–10 somites). Embryos are viewed laterally with anterior to the top to examine KV expression. b, d, f, h, j, l Results of in situ hybridization of candidate genes and standard control at 24 hpf (primordium 5 stage). Lateral view of embryos with anterior to the left. KV (blue arrow), floor plate (red arrows), pronephric duct (green arrows), notochord (yellow arrow), head (black arrows), ubiquitous expression (orange arrows) EXPRESSION / LABELING:
|
|
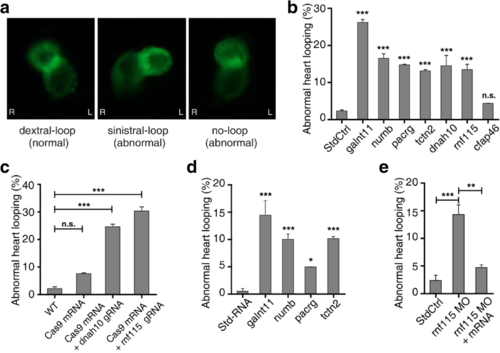
Loss of function of candidate genes in zebrafish disturbed cardiac looping. a Zebrafish heart shows normal dextral loop, abnormal sinistral loop, and no-loop types in cmlc2:eGFP morphants in ventral view. b The percentage of abnormal heart looping with MO injected. The experiments were repeated 3 times, and at each time > 70 embryos were examined for each group. c Summary of the abnormal heart looping of dnah10 and rnf115 mutations generated by co-injection of zebrafish Cas9 mRNA 600 pg and dnah10 gRNA 100 pg or rnf115 gRNA 100 pg. The experiments were repeated 3 times, and at each time > 71 embryos were examined for each group. d Percentage of embryos that exhibit abnormal cardiac looping with mRNA over-expressed. e The rnf115 mRNA can rescue LR randomization. The abnormal heart looping phenotype which is induced by rnf115 MO can be rescued by 6.25 pg rnf115 mRNA. Heart looping direction was assayed in zebrafish at stage 2 dpf. Bars show the total percent of abnormally looped heart including two types: no-loop and sinistral loop heart. Standard control MO (StdCtrl) is negative control. galnt11 is used as positive control. Error bars represent the standard error of the mean (SEM). *P < 0.05, **P < 0.01, ***P < 0.001, respectively vs. StdCtrl. WT, wild type PHENOTYPE:
|
|
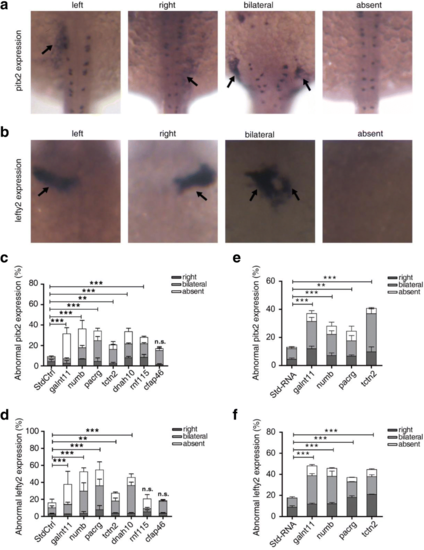
Analysis of pitx2 and lefty2 expression patterns in the lateral plate mesoderm in 18–22 somites. a The expression of pitx2 exhibits four patterns, left, right, bilateral, or absent, in the posterior lateral plate mesoderm of zebrafish embryos. b Morphants show left, right, bilateral or absent lefty2 expression in the cardiac field. c, d Summary of abnormal pitx2 and lefty2 mRNA expression in zebrafish morphants. e, f Summary of abnormal pitx2 and lefty2 mRNA expression in zebrafish with mRNA over-expressed. Embryos are viewed dorsally with anterior to the top. Bars show the percent of abnormal pitx2 and lefty2 expression including three types: right, bilateral, absent expression. Standard control (StdCtrl or Std-RNA) is negative control. galnt11 is used as positive control. Error bars represent the SEM. *P < 0.05, **P < 0.01, ***P < 0.001, respectively, vs. standard control EXPRESSION / LABELING:
PHENOTYPE:
|