- Title
-
Temporal control over the initiation of cell motility by a regulator of G-protein signaling
- Authors
- Hartwig, J., Tarbashevich, K., Seggewiß, J., Stehling, M., Bandemer, J., Grimaldi, C., Paksa, A., Groß-Thebing, T., Meyen, D., Raz, E.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
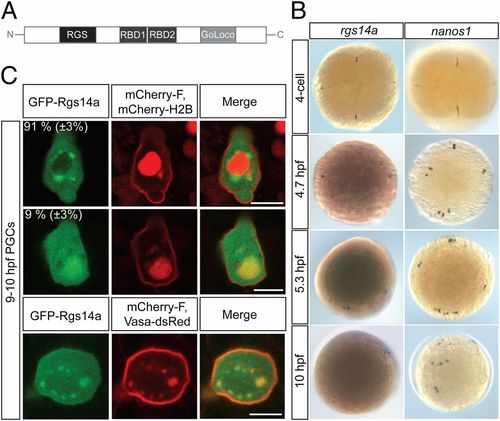
Zebrafish regulator of G-protein signaling 14a (rgs14a) RNA expression and Rgs14a protein localization. (A) A schematic representation of the Rgs14a protein structure showing the N-terminal RGS domain, the two Raf-like Ras-binding domains (RBD), and the C-terminal GoLoco domain. (B) Whole-mount in situ RNA hybridization using rgs14a (Left) and nanos (Right) antisense RNA probes at the indicated embryonic stages; 5.3- and 10-hpf embryos were overstained in the case of rgs14a in situ hybridization to detect the weak expression in the PGCs. (C) GFP-Rgs14a fusion protein expressed in PGCs is localized to the cytoplasm and the plasma membrane, with enrichment in perinuclear granules (in 91% of the 46 PGCs analyzed; Top) or in the nucleus (in 9% of the 46 PGCs analyzed; Middle). The perinuclear granules where the GFP-Rgs14a protein is localized harbor Vasa-DsRed protein (Bottom), defining them as germ cell granules. (Scale bars, 10 μm.) |
|
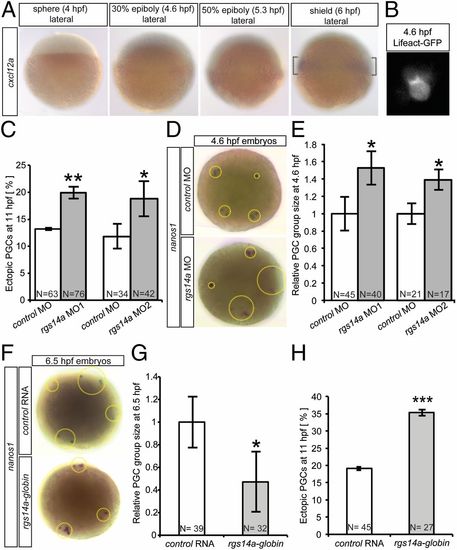
The role of Rgs14a in the temporal control of migration initiation. (A) Whole-mount in situ hybridization of embryos with a cxcl12a antisense RNA probe showing the initiation of transcription of the chemokine between 5.3 and 6.0 hpf. (B) Premigratory PGCs develop polarized actin brushes. (C) Morpholino-mediated knockdown of rgs14a leads to an increase in the average number of ectopic PGCs at 11 hpf (number of ectopic PGCs divided by total PGC number; Fig. S3). (D and E) PGC groups at 4.6 hpf (dotted circles) in Rgs14a knockdown embryos cover 53% larger areas than those in control embryos. (F and G) Early overexpression of Rgs14a, using the Xenopus globin32UTR, leads to significantly smaller PGC group areas at 6.5 hpf and an increase in the average number of ectopic PGCs at 11 hpf; target defined analogous to Fig. S3 (H). PGC groups in D–G were labeled with a nanos1 antisense RNA probe. Control embryos in F–H expressed a pa-(photoactivatable)-gfp-globin32UTR mRNA. For E and G, PGC group sizes were averaged for each embryo, and 32–45 embryos were included per experimental point. n = number of embryos analyzed. (C and H) *P < 0.05, **P < 0.01, and ***P < 0.001 using Student t test. (E and G) *P < 0.05 using Mann–Whitney u test. Error bars indicate SEM. EXPRESSION / LABELING:
PHENOTYPE:
|
|
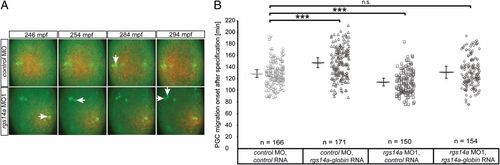
The acquisition of cell motility is inhibited by Rgs14a. (A) Snapshots from time-lapse movies showing PGC migration-onset events (mpf = minutes post fertilization). Rgs14a morphants (MO1) exhibit earlier PGC migration onset events (white arrows) compared with control morphants (Movie S1). (B) PGC migration onset events for Rgs14a and control morphants, as well as Rgs14a overexpression. Knockdown of Rgs14a leads to premature migration onset, whereas overexpression of 100 pg rgs14a-globin32UTR delays PGC migration onset. Combination of both treatments restored WT motility acquisition timing. Time point zero is defined as the PGC specification event at 180 mpf (9). pa(photoactivatable)-gfp-globin32UTR was used as a control mRNA. n = number of cells analyzed. (B) ***P < 0.001 using Mann–Whitney u test. Error bars indicate SEM. |
|
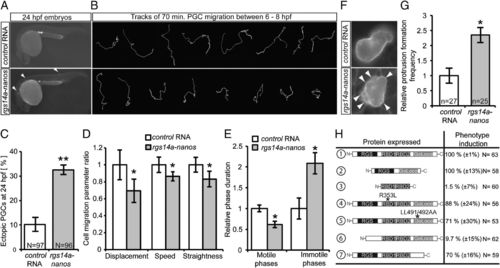
Expression of Rgs14a during migration phases impairs PGC motility. (A and C) Forced expression of rgs14a-nanos32UTR in PGCs during migratory stages results in inefficient colonization of the gonad region at 24 hpf. Ectopic cells exemplarily labeled with arrowheads in A, n = number of embryos analyzed. (B) PGCs expressing Rgs14a exhibit shorter migration tracks, stemming from impaired motility parameters (D) and longer immotile phases (E). The results presented in B, D, and E were derived from time-lapse movies spanning 70 min (Movies S3 and S4) captured in three independent experiments, with 21 control and 35 rgs14a expressing PGCs analyzed. (F and G) PGCs expressing Rgs14a display an apolar cell shape and extensive protrusion formation (protrusions indicated by white arrowheads) (Movies S5 and S6), n = number of cells analyzed. (H) The RGS domain of Rgs14a is essential for the induction of ectopic PGC migration when expressed in PGCs under the control of the nanos 32 UTR. n = number of embryos analyzed. A, C, F, G, and H were controlled by injection of cd14-nanos32UTR RNA. B, D, and E were controlled by injection of gfp-nanos32UTR RNA. *P < 0.05 and **P < 0.01 using Student t test. Error bars indicate SEM. |
|
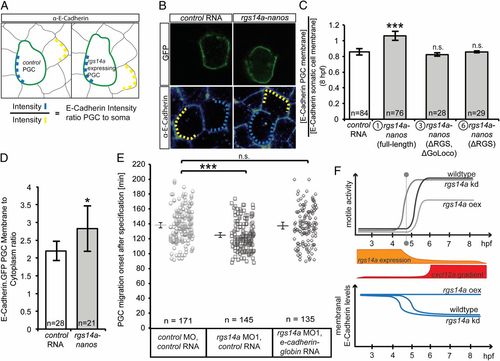
Rgs14a elevates E-cadherin levels on PGC membranes thereby affecting PGC motility. (A) The experimental setup and the scheme for evaluating E-cadherin level. (A and B) Embryos expressing F-EGFP on their membranes were injected with rgs14a-nanos3′UTR and control mRNA (cd14-nanos3′UTR), fixed, and stained for E-cadherin at 8 hpf. E-cadherin signal on the membrane of PGCs (light blue dashed lines) was normalized by dividing it by the corresponding signal on the membranes of neighboring somatic cells (yellow dashed lines). (C) PGCs expressing the full-length Rgs14a (protein 1) during migratory stages exhibit more E-cadherin on their membrane compared with PGCs expressing control RNA or RNA encoding for versions 3 and 6 of the Rgs14a protein. n = number of cell pairs analyzed. (D) Expression of Rgs14a in PGCs enhances the membrane localization of E-cadherin-GFP relative to the cytoplasm. (E) Forced E-cadherin expression abrogates the premature motility acquisition of MO1-treated PGCs. (F) Summary of the findings. The initiation of spatially restricted cxcl12a expression (red) coincides with a rapid decrease in rgs14a mRNA levels in PGCs (orange), suggesting a role in PGC migration onset (vertical dotted line). The onset of motile activity of PGCs occurs prematurely in germ cells knocked down (kd) for rgs14a, whereas the overexpression (oex) of Rgs14a led to motility defects, correlated with enhanced E-cadherin levels in the cells, whereas rgs14a knockdown prematurely reduced E-cadherin levels. n = number of cells analyzed. (C and E) ***P < 0.001 using Mann–Whitney u test. (D) *P < 0.05 using Student t test. Error bars indicate SEM. EXPRESSION / LABELING:
|
|
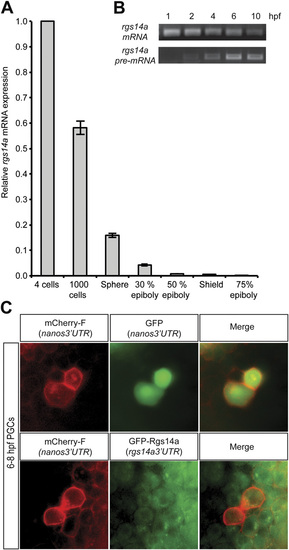
(A) qPCR analysis of rgs14a mRNA showed gradual decrease of maternally provided RNA between the four-cell and 75% epiboly stages. (B) Performing RT-PCR for either total rgs14a mRNA or specifically for the unspliced pre-mRNA pool reveals that the gene is zygotically expressed. (C) Injection of mRNA encoding for mCherry cloned upstream of the nanos3′UTR together with mRNA encoding for GFP cloned upstream of the nanos3′UTR results in enriched expression of both fluorescent proteins in the PGCs (Upper). In contrast, injection of mRNA encoding for mCherry cloned upstream of the nanos3′UTR together with mRNA encoding for GFP fused to the ORF of the Rgs14a protein cloned upstream of the rgs14a3′UTR results in uniform GFP expression in all cells of the embryo (Lower). |