- Title
-
Dvr1 transfers left-right asymmetric signals from Kupffer's vesicle to lateral plate mesoderm in zebrafish
- Authors
- Peterson, A.G., Wang, X., and Yost, H. J.
- Source
- Full text @ Dev. Biol.
|
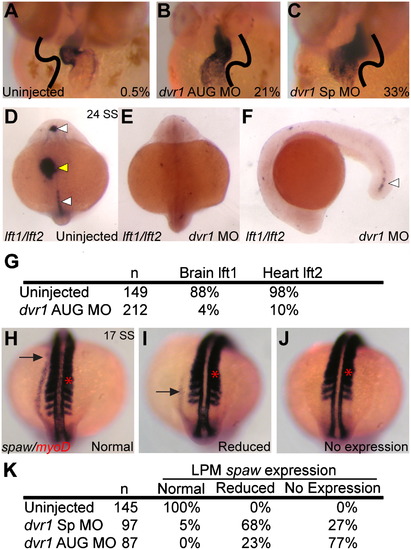
Knockdown of dvr1 in zebrafish causes heart, brain and LPM laterality defects. (A)–(C) Heart looping in control and morphant embryos, ventral view, cmlc2 in situ hybridization. Value in each panel indicates the percentage of reversed heart looping, orientation indicated by black lines. (A) Uninjected controls. (B) dvr1 AUG morphants. (C) dvr1 Sp morphants. (D)–(F) Asymmetric expression of lft1 in brain and expression in midline (white arrowheads) and asymmetric expression in lft2 in heart (yellow arrowhead) in control embryos (D) is lost in dvr1 morphants at 24 SS stage (E). Only a remnant lft1 expression is maintained in posterior midline (F). (G) Table showing quantification of brain lft1 and heart lft2 expression in both uninjected and dvr1 AUG morphant embryos. Values indicate percentage of normal expression. (H)–(J) Examples of different categories of spaw lateral plate mesoderm expression (black arrow) at 17 SS stage. myoD expression is also shown as a reference for spaw migration (red arrow). (H) Normal left-sided expression pattern of spaw. (I) Reduced spaw expression. (J) No spaw expression. (K) Quantification of spaw expression. In contrast to left-sided expression in control embryos, morphant embryos had reduced or no spaw expression in LPM. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Early expression patterns of dvr1 mRNA. Maternal dvr1 mRNA was detected from (A) 4-cell stage to (B) 50% epiboly. (C) Zygotic dvr1 expression at tailbud stage, in LPM and tailbud. Green bars indicate regions for cross sections in (D) and (E), showing dvr1 expression in LPM and tailbud. (F) Dorso-anterior view of dvr1 expression in the heart field at the 17 SS stage. (G) Posterior view at 17 SS stage, showing persistent tailbud expression and no expression in poserior LPM. EXPRESSION / LABELING:
|
|
Initiation in LPM can occur in dvr1 morphants and timing is dependent on mix of activators and inhibitors. (A) Control (BSA injected) wildtype embryos show normal left-sided spaw expression. Embryos injected with Nodal protein on right side, (B) anterior view and (C) same embryo, posterior view, had both native left-sided and ectopic right-sided LPM spaw expression (black arrows). Midline lft1 expression (white arrow) and somitic myoD expression serve as spatial landmarks. (D) Control (BSA injected) dvr1 morphant embryos exhibit no spaw expression. (E) and (F) dvr1 morphant embryos injected with Nodal protein on the right side exhibit ectopic right-sided spaw expression. (E) Same Nodal-injected dvr1 morphant, (E) posterior and (F) anterior views. (G) Quantification of right-sided spaw expression in uninjected controls, BSA injected controls, Nodal protein injected into right side of wildtype embryos, dvr1 morphants injected with BSA on the right side, and dvr1 morphants injected with Nodal protein on the right side. (H) Quantification of left-sided (normal) spaw expression in single and double morpholino injected embryos. (I) Quantitative measurements of initiation and posterior-to-anterior propagation of spaw expression in LPM of wildtype, cha morphants, and dvr1+cha double morphants. |
|
KV morphology and flow is normal in dvr1 morphants. (A) and (B) Cha expression in (A) uninjected embryos and (B) dvr1 morphants. (C) and (D) Peri-nodal spaw expression in (C) uninjected and (D) dvr1 morphant embryos. (E) and (F) KV shape in (E) uninjected and (F) dvr1 morphant embryos. (G) KV cilia number in wildtype and dvr1 morphant embryos. (H) and (I) KV cilia labeled by α-alpha-tubulin antibody in (G) uninjected and (H) dvr1 morphant embryos. (J) KV cilia length in wildtype and dvr1 morphant embryos. (K) and (L) Fluorescent bead tracking of fluid flow in KV in (K) uninjected wildtype and (L) dvr1 morphants. Each colored line represents the track of an individual bead. (M) Average fluorescent bead velocity. EXPRESSION / LABELING:
PHENOTYPE:
|
|
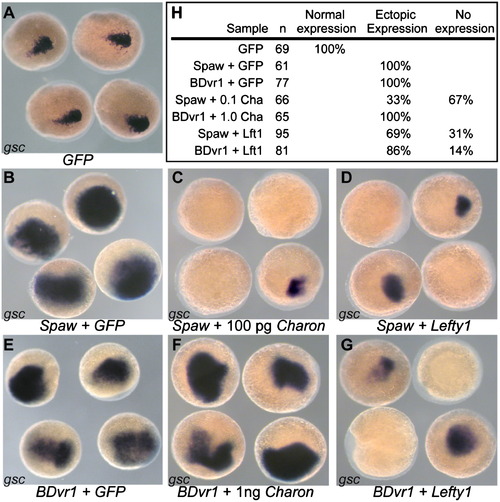
Charon inhibits Spaw but not BDvr1. (A) Endogenous gsc expression in control GFP mRNA injected zebrafish embryos. (B) spaw mRNA alone expands the gsc expression domain while co-injection of either (C) charon mRNA or (D) lefty1 mRNA suppresses the gsc expression enhancement caused by spaw alone. (E) Injection of Bdvr1 mRNA expands the expression domain of gsc. (F) Coinjection of charon with Bdvr1 cannot block the ectopic expression of gsc induced by Bdvr1 even when charon mRNA is injected at 10 times the concentration as that able to suppress ectopic gsc expression caused by spaw. (G). Lefty1 coinjection with Bdvr1 is able to suppress the ectopic gsc expression caused by Bdvr1. (H) Quantification of gsc expression patterns in injected embryos. |
|
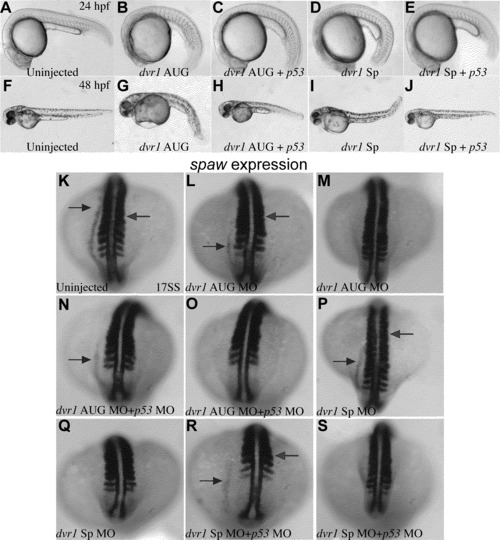
Dvr1 LR phenotype is not rescued by p53 morpholino. (A)–(E) At 24, Dvr1 AUG and Sp morphants (B) and (C) show a head necrosis phenotype that can be rescued by coinjection of p53 morpholino suggesting that this phenotype is nonspecific. (F)–(J) At 48 hpf nonspecific morpholino phenotypes in both Dvr1 AUG and Sp morphants (G) and (I) can also be rescued by p53 morpholino co-injection. (K)–(S) The reduced or absent expression of the left-sided LPM marker, spaw, in Dvr1 AUG and Sp morphants is a Dvr1 morpholino specific phenotype and cannot be rescued by co-injection with p53 morpholino co-injection. |
|
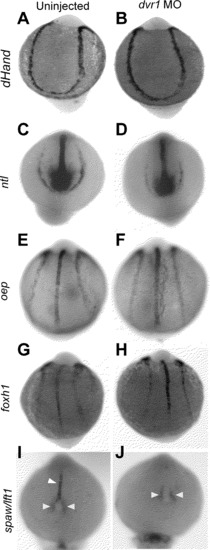
LPM Markers and Midline Markers are Normal in Dvr1 Mo (A) and (B) hand2 expression. (C) and (D) ntl expression. (E) and (F) oep expression. (G) and (H) foxh1 expression. (I) and (J) lft1/spaw expression. White arrowhead indicate lft1 expression and yellow arrowheads indicate peri-KV spaw expression. All embryos are about 10 SS. (A,C,E,G, I) uninjected embryos. (B,D,F,H,J) are dvr1 AUG MO. All images are dorsal view with anterior up. |
Reprinted from Developmental Biology, 382(1), Peterson, A.G., Wang, X., and Yost, H. J., Dvr1 transfers left-right asymmetric signals from Kupffer's vesicle to lateral plate mesoderm in zebrafish, 198-208, Copyright (2013) with permission from Elsevier. Full text @ Dev. Biol.