- Title
-
miR-153 Regulates SNAP-25, Synaptic Transmission, and Neuronal Development
- Authors
- Wei, C., Thatcher, E.J., Olena, A.F., Cha, D.J., Perdigoto, A.L., Marshall, A.F., Carter, B.D., Broadie, K., and Patton, J.G.
- Source
- Full text @ PLoS One
|
miR-153 regulates embryonic movement. Embryonic movement was recorded at 1 dpf for each of the singly and multiple injected conditions shown (see Movies). The number of twitches per minute was counted and significance determined by comparing the noninjected control (NIC) embryos to all other conditions using ANOVA with Dunnett’s post-test. *, p<0.05; **, p<0.01. Movements were counted for approximately 60 embryos over 2–5 minutes for each condition. PHENOTYPE:
|
|
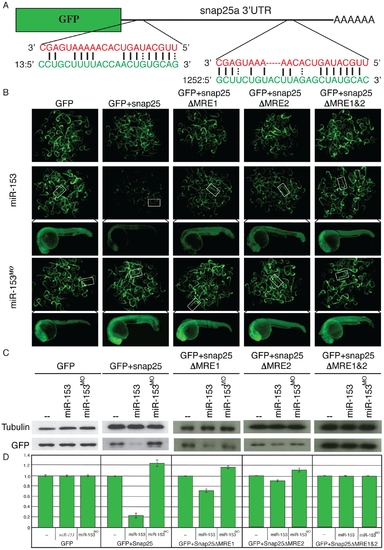
miR-153 targets snap-25a. (A) GFP reporter constructs were created by fusing the reading frame of GFP to the snap-25a 32UTR. Two predicted miRNA recognition elements (MREs) were identified in the snap-25a 32 UTR. The miR-153 sequence is indicated in red and the corresponding snap-25a UTR sequence is shown in green. (B) Single cell zebrafish embryos were injected with mRNAs derived from GFP reporters lacking a UTR (GFP), fused to the full length snap-25a UTR (+snap-25), or mutant versions of the snap-25a UTR lacking individual MREs (snap-25aΔMRE1 and snap-25a”MRE2) or both MREs (snap-25aΔMRE1&2). Embryos were injected in the presence or absence of exogenous miR-153 or morpholinos against miR-153 (miR-153MO). Fluorescence levels were examined at 1 dpf. Clusters of embryos (~60) are shown as well as a high magnification image of a single representative embryo. (C) Lysates from ~100 embryos were prepared from embryos treated as in B and GFP protein levels were determined by western blotting using antibodies against GFP or control antibodies against α-tubulin. (D) Quantitation of westerns was performed with a paired Student’s t-test (n = 5). |
|
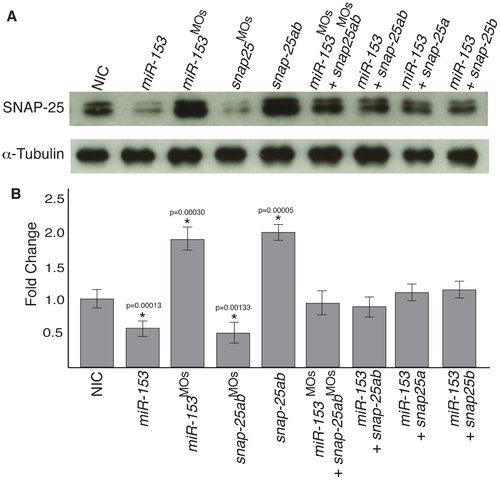
miR-153 regulates endogenous snap-25a expression. (A) Embryo lysates were prepared from either NIC embryos or embryos injected with miR-153, miR-153MO, mRNAs encoding snap-25a and snap-25b, morpholinos against snap-25, or combinations thereof, as indicated. Western blots were performed using antibodies against SNAP-25 and α–tubulin. (B) Quantification of SNAP-25 levels from the western blots (n = 3) shown in A. Significance was determined by a two-tailed Student’s t-test. Error bars show s.e.m. |
|
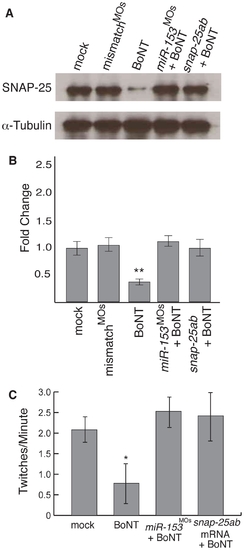
miR-153 mimics the effects of BoNT A. (A) Single cell embryos were injected as indicated and then at 27 hpf, exposed to Botulinum neurotoxin A (BoNT) for 30 minutes. After recovery for 1 hour, western blots were performed on embryo lysates using antibodies against SNAP-25 or α–tubulin. (B) Quantitation of SNAP-25 levels from A, n = 3. **, p<0.01 (C) Embryonic movement in the presence or absence of BoNT A. The number of twitches per minute was counted as in Fig. 1 for embryos treated as indicated. Significance was determined by comparing mock embryos to all other conditions using ANOVA with Dunnett’s post-test, n = 15. *, p<0.05. |
|
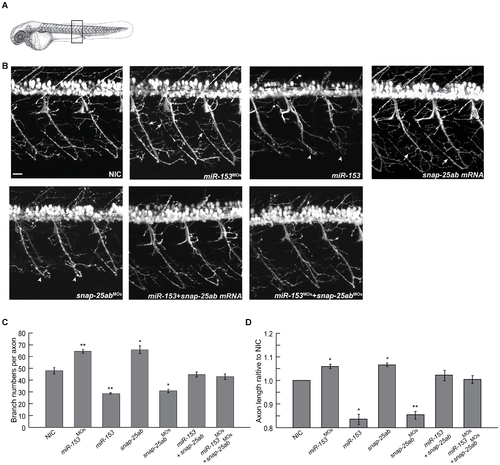
miR-153 regulates the morphology and structure of motor neurons. (A) A transgenic zebrafish line, Tg(mnx1:TagRFP-T), that expresses RFP in motor neurons was used to monitor the effects of altered levels of miR-153 and snap-25 at 55 hpf. For all confocal images, developing motor neurons were examined from the same somites, as indicated. (B) Morphology of developing motor neurons under each of the indicated conditions. Arrows indicate increased branching after knockdown miR-153 (miR-153MO) or overexpression snap-25a,b mRNA. Arrowheads indicate the structural defects after miR-153 overexpression or knockdown of snap-25a,b (snap-25a,bMO). Scale bar: 20 µm. (C) Quantification of motor neuron axonal branch number under the different conditions shown in (B). Error bars show s.e.m. Significance was determined using ANOVA with Dunnett’s post-test, n = 5. *, p<0.01; **, p<0.005. (D) Quantification of motor neuron axon length relative to uninjected control under the different conditions shown in (B). Error bars show s.e.m. ANOVA with Dunnett’s post-test, n = 5. *, p<0.05; **, p<0.01. |
|
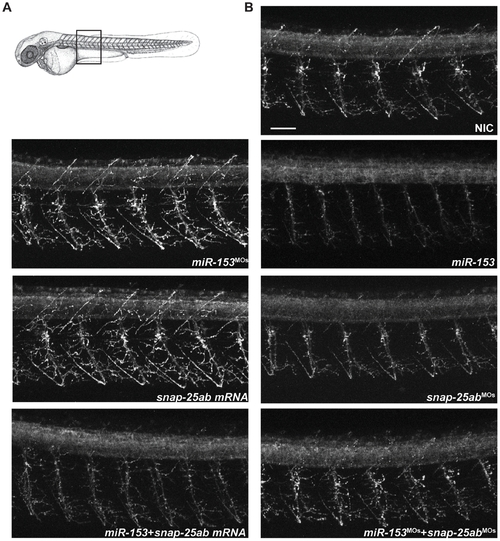
miR-153 regulates primary motor neuron development. (A) Immunofluorescence performed on whole mount zebrafish embryos at 55 hpf using Znp-1 antibodies to label primary motor neurons. Confocal images were acquired from the same somites for all embryos, as indicated. (B) Effects on primary motor neuron structure and branching under the indicated conditions. Scale bar: 40 μm. |
|
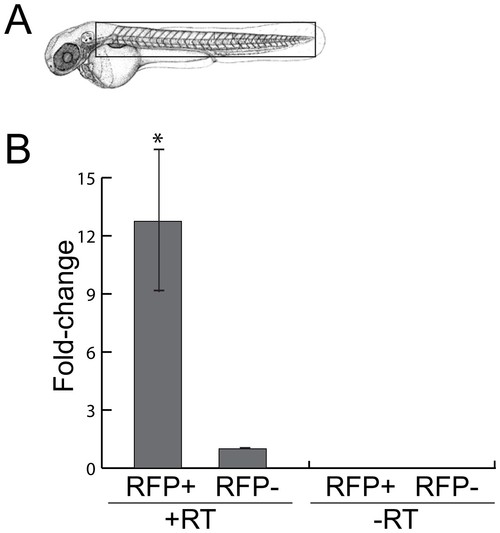
miR-153 is expressed in motor neurons. To enrich for motor neurons, heads were removed from 52 hpf embryos just posterior to the otic vesicle and trunks were dissociated to facilitate sorting of RFP+ and RFP- cells. RNA was isolated from these cell fractions and RT/PCR was performed to determine miR-153 levels relative to U6 snRNA. Significance was determined by a two-tailed Student’s t-test with the error bars representing s.e.m.; p<0.02. EXPRESSION / LABELING:
|
|
miR-153 regulates synaptic activity at the neuromuscular junction. (A) FM1-43 loading of neuromuscular junction (NMJ) boutons in 55 hpf fish embryos. (B) Postsynaptic clusters of AChRs were labeled with Alexa 594-conjugated α-bungarotoxin. Overexpression of miR-153 caused decreased FM1-43 loading, indicating down-regulation of the synaptic vesicle cycle within NMJ boutons (arrowheads). (C) Knockdown of miR-153 (miR-153MO) promoted greater uptake of FM1-43 dye, indicating increased synaptic vesicle cycling. Scale bar: 10 μm. (D) Quantification of FM1-43 fluorescent intensity with a paired Student’s t-test. Error bars show s.e.m. *p<0.01; **p<0.02. |
|
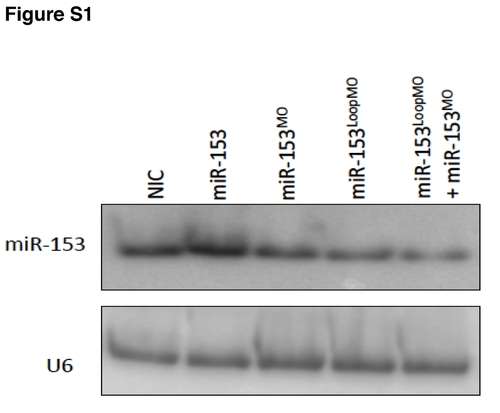
Northern blot of miR-153 overexpression and knockdown. Perturbation of miR-153 expression levels by injection of miR-153 or MOs against different regions of pre-miR-153 was verified by northern blot. U6 served as a loading control. |
|
miR-153 targets snap-25b. (A) GFP reporter constructs were created by fusing the reading frame of GFP to the snap-25b 32UTR. Three predicted miRNA recognition elements (MREs) were identified in the snap-25b 32 UTR. The miR-153 sequence is indicated in red and the corresponding snap-25a UTR sequence is shown in green. (B) Single cell zebrafish embryos were injected with mRNAs derived from GFP reporters lacking a UTR (GFP), fused to the full length snap-25b UTR (GFP+snap-25b), or mutant version of the snap-25b UTR lacking all MREs (GFP+snap-25bΔMRE1, 2&3). Embryos were injected in the presence or absence of exogenous miR-153 or morpholinos against miR-153 (miR-153MO). Fluorescence levels were examined at 1 dpf. Clusters of embryos (~30) are shown. (C)Lysates from ~100 embryos were prepared from embryos treated as in B and GFP protein levels were determined by western blotting using antibodies against GFP or control antibodies against α-tubulin. |
|
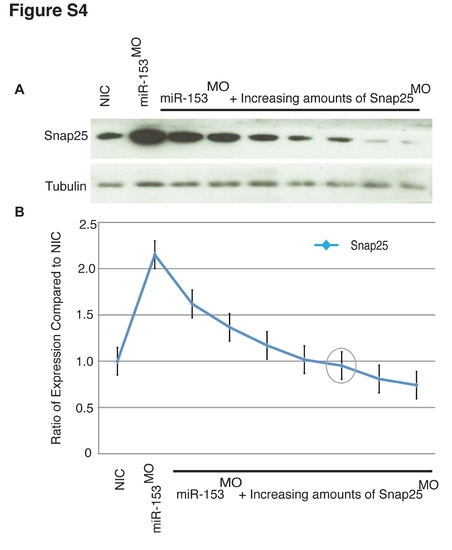
Dose-dependent rescue of miR-153 knockdown. (A) Single cell embryos were injected with a constant level of miR-153MO and increasing amounts (increments of 2 ng) of snap-25MOs. Embryo lysates from ~60 embryos in each group were prepared and SNAP-25 protein levels determined by western blotting. (B) Quantitation of westerns (n = 3) from A. The grey circle represents the amount of snap- 25MO (10 ng) used in co-injection rescue experiments. |
|
Dose-dependent rescue of miR-153 over-expression. (A) Single cell embryos were injected with a constant level of miR-153 and increasing amounts (increments of 50 pg) of snap-25a, snap-25b, or snap-25a&b mRNA. Embryo lysates from ~60 embryos were prepared from embryos in each treatment group and SNAP-25 protein levels were determined by western blotting. (B) Quantitation of westerns (n = 3) from A. The grey circles represent the amounts used in co-injection rescue experiments (75 pg each of snap-25a and b, 250 pg of snap-25a, and 300 pg of snap-25b). |
|
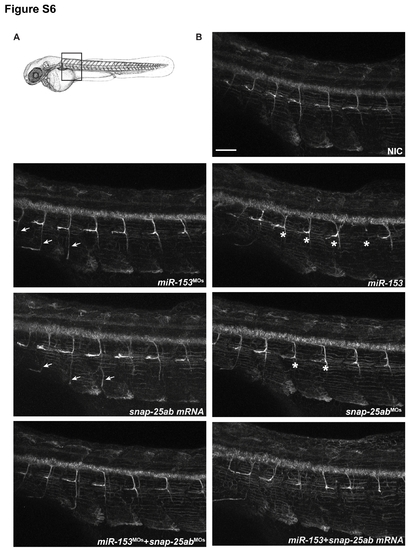
miR-153 regulates secondary motor neuron development. (A) Immunofluorescence was performed on whole mount zebrafish embryos at 55 hpf using Zn-8 antibodies to label secondary motor neurons. Confocal images were acquired from the same somites for all embryos, as indicated. (B) miR-153 knockdown (miR-153MO) and snap-25a,b overexpression significantly increased the growth of secondary motor neuron axons (arrows). Overexpression of miR-153 or knockdown of snap-25a,b (snap-25a,bMO) caused severe defects in axon development and architecture (asterisks). Scale bar: 40 μm. |