- Title
-
Distinct requirements for wnt9a and irf6 in extension and integration mechanisms during zebrafish palate morphogenesis
- Authors
- Dougherty, M., Kamel, G., Grimaldi, M., Gfrerer, L., Shubinets, V., Ethier, R., Hickey, G., Cornell, R.A., and Liao, E.C.
- Source
- Full text @ Development
|
Palate morphogenesis in zebrafish. (A,B) The anterior cells at 14-somites (arrowhead, A) tracked to the median ethmoid plate at 4.5 dpf (arrowheads, B). (C) Screenshots of time-lapse movies capturing this process. (D,E) Unilateral labeling of the entire maxillary prominence at 55 hpf (arrowhead, D) revealed that the mature lateral ethmoid plate and trabeculae are formed both by uniform expansion (bracket, E) as well as increased proliferation at the leading edge (asterisk) (see also Fig. 2C, supplementary material Movie 4). (F,G) Photoconversion of FNP cells at 20 somites (F arrowhead) labeled only the median ethmoid at 4.5 dpf (G), also captured in supplementary material Movies 1-3. (H) Enlargement of inset in G. Inspection of the ethmoid plate revealed that labeled CNCCs could be traced along the seam (broken line) between the median and lateral ethmoid plate; lateral to this line, chondrocytes are columnar in appearance while medially they are cuboidal. A, dorsal view; F, lateral view; B,D,E,G,H, ventral views. Scale bar: 50 μm. EXPRESSION / LABELING:
|
|
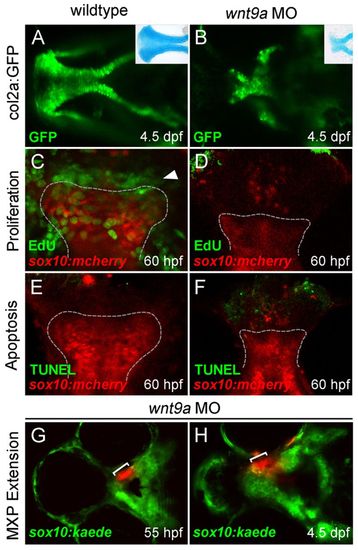
wnt9a knockdown results in failure of maxillary extension. (A,B) wnt9a knockdown in the col2a:GFP transgenic (B) produced an ethmoid plate similar to that seen by Alcian Blue stain (inset, B). (C,D) EdU staining from 55-60 hpf revealed a proliferative front in the leading edge of the extending ethmoid plate during palatogenesis (arrowhead, C), but absence of new cell divisions in morphants (D). (E,F) TUNEL assay shows the shortened ethmoid plate (outlined) is not a result of increased cell death. (G,H) When maxillary process CNCCs were labeled by kaede photoconversion before maxillary extension (bracket, G), they failed to extend significantly by 4.5 dpf (H). All images are ventral views, anterior leftwards. |
|
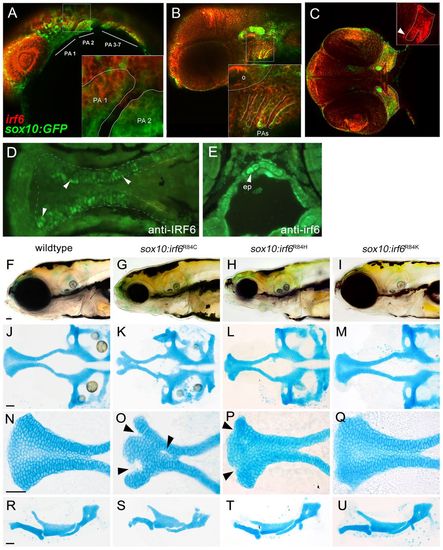
Expression analysis of endogenous irf6 and functional analysis of sox10:irf6R84C,H,K mutants. (A-E) RNA in situ hybridization demonstrated extensive overlap of endogenous irf6 expression with sox10-labeled CNCCs. (A) At 24 hpf, irf6 expression can be found pharyngeal arches. (B) By 48 hpf, irf6 expression is detected in both the pharyngeal mesenchyme and pharyngeal ectoderm. (C) By 55 hpf, irf6 expression is detected in the converging maxillary and mandibular prominences (inset). (D,E) Immunohistochemistry at 72 hpf using antisera against non-overlapping epitope of human IRF6 (D) and zebrafish irf6 (E), each with specific staining in the ethmoid plate, shown in ventral and cross-sectional views. Arrowheads indicate ethmoid plate chondrocytes. (F-Q) Comparison of wild-type and mutant craniofacial structures, with live images (F-I), dissected neurocranium (J-M), flat-mounted lower jaw structures (R-U) at 10× magnification and ethmoid plate (N-Q) at 40× magnification. The R84C cleft phenotype is the most severe (arrowheads, O). R84H has a milder cleft phenotype, with subtle indentations at the anterior edge of the seam between median and lateral ethmoid (arrowheads, P), whereas the conserved amino acid substitution R84K appears wild type. ep, ethmoid plate; PA, pharyngeal arch; o, otic vesicle. EXPRESSION / LABELING:
PHENOTYPE:
|
|
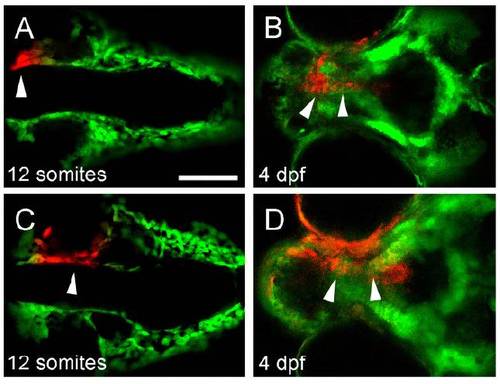
Lineage analysis of cranial neural crest in wnt9a morphant. (A-D) CNCCs anterior to the eye, which are destined to form the median ethmoid plate (arrow, A), still migrate to the median region of the dysmorphic and foreshortened ethmoid plate (arrows, B), whereas cells medial to the eye, which are destined to form the lateral ethmoid plate (arrow, C) also reach their destination at 4.5 dpf (arrows, D), indicating that the shortened ethmoid plate is not a result of failure of early CNCC migration. Scale bar: 50 μm. |
|
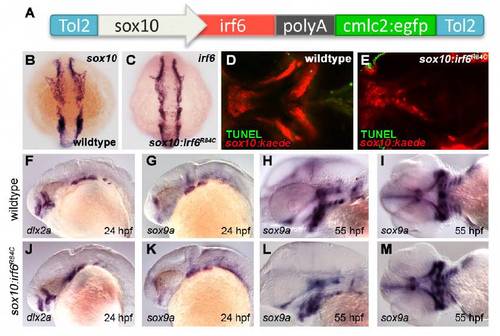
Characterization of sox10:irf6R84C transgenics. (A) Transgene contains mutant irf6 (R84C, R84H, or R84K) under the control of the sox10 promoter, followed by a fluorescent tag (cmlc2:egfp) to facilitate screening. (B,C) Expression pattern of exogenous irf6 matches that of sox10 at 10 somites. (D,E) TUNEL assay for apoptosis confirms that clefting in sox10:irf6R84C is not a result of cell death. (F,G,J,K) Expression analysis of neural crest markers dlx2a and sox9a show that early neural crest development is not significantly affected in the irf6 mutant. (F,J) At 24 hpf, the expression pattern of dlx2a is unaffected in the developing pharyngeal arches. (G,K) Similarly, pharyngeal arch expression of sox9a is similar in wild-type and mutant embryos. (H,I,L,M) By 55 hpf, although the head is already noticeably smaller in mutants, patterning of sox9a is similar, highlighting the developing ethmoid plate, mandible, ceratohyal and ceratobranchials. |