- Title
-
The feelgood mutation in zebrafish dysregulates COPII-dependent secretion of select extracellular matrix proteins in skeletal morphogenesis
- Authors
- Melville, D.B., Montero-Balaguer, M., Levic, D.S., Bradley, K., Smith, J.R., Hatzopoulos, A.K., and Knapik, E.W.
- Source
- Full text @ Dis. Model. Mech.
|
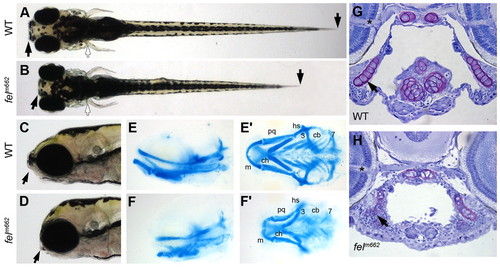
The feelgood mutation affects craniofacial skeletal development. (A–D) Live images of wild-type (WT) and feelgood (felm662) embryos at 5 dpf. Arrows indicate reduced length of head and trunk in dorsal (A,B) and lateral (C,D) views. (E–F′) Alcian blue staining of cartilage elements in head skeleton in lateral (E,F) and ventral (E′,F′) views. cb, ceratobranchials 3–7; ch, ceratohyal; hs, hyosymplectic; m, Meckel’s cartilage; pq, palatoquadrate. (G,H) Toluidine blue staining of transverse sections of the jaw at the level of the optic nerve (asterisk). Nuclei stain blue, whereas ECM stains purple. Arrow indicates tightly packed nuclei in feelgood mutants. PHENOTYPE:
|
|
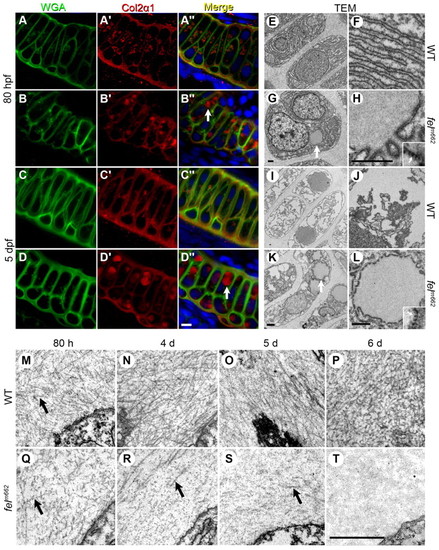
Protein trafficking is disrupted in feelgood mutants. (A–D′′) Immunostaining of WGA (A–D) and Col2α1 (A2–D′) in the Meckel’s cartilage of 80-hpf (A–B′′) and 5-dpf (C–D′′) wild-type (WT; A-A′′, C-C′′) and feelgood. (B-B3, D-D3) embryos. Arrows in merged images indicate aberrant intracellular collagen localization in feelgood. Scale bar: 5 μm. (E–L) TEM images of 80-hpf (E–H) and 5-dpf (I–L) WT and feelgood chondrocytes. Arrows indicate distended ER membranes in feelgood cells. (M–T) TEM images of collagen fibrils in the extracellular space of feelgood mutants and WT siblings at 80 hpf and 4, 5 and 6 dpf. Arrows point to representative individual collagen fibrils. Scale bar: 1 μm. PHENOTYPE:
|
|
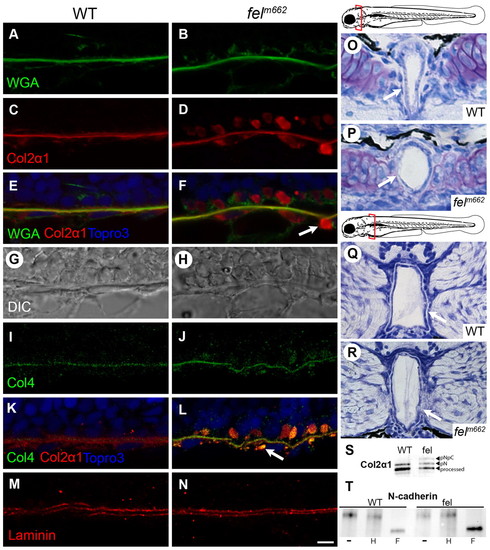
Collagen trafficking is preferentially disrupted in feelgood mutants, leading to notochord defects. (A–F) Immunofluorescence of WGA, Col2α1 and merged images of 15-µm sagittal sections of the notochord of 28-hpf embryos. Arrow indicates vesicle-like collagen staining outside the notochord sheath. (G,H) DIC images of the corresponding sections in A–F. (I–N) Immunofluorescence of type-IV and type-II collagen (I–L) and laminin (M,N) in 15-μm sagittal sections of the notochord at 28 hpf. Scale bar: 5 μm. (O–R) Toluidine blue staining of transverse sections through the notochord at the level of the posterior parachordal plate (O,P) and posterior medulla oblongata (Q,R) in 80-hpf embryos. Schematic diagrams mark the plane of the corresponding sections. Arrows indicate the less robust notochord sheath in feelgood compared with wild types. (S) Immunoblot analysis of type II collagen processing. Molecular forms are indicated as: processed, the fully processed form; pN, pN-collagen II; pNpC, unprocessed procollagen II. (T) EndoH sensitivity assay for N-cadherin. Embryo lysates were either untreated (–) or treated with either EndoH (H) or PNGase F (F) before immunoblotting for N-cadherin. PHENOTYPE:
|
|
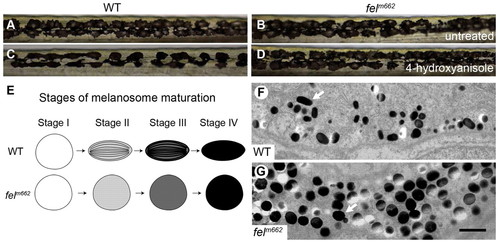
Melanophore development is disrupted in feelgood mutants. (A–D) Live image of melanophores in the trunk of 6-dpf wild-type (WT) and feelgood mutants in untreated (A,B) and 0.5 μg/ml 4-hydroxyanisole treated (C,D) embryos. (E) Summary drawing of the stages of melanosome maturation comparing normal and feelgood melanocytes. (F,G) TEM images of 5-dpf melanosomes. Arrows point to a mature melanosome in WT (F), and a round, dark, stage IV melanosome in feelgood (G). Scale bar: 1 μm. PHENOTYPE:
|
|
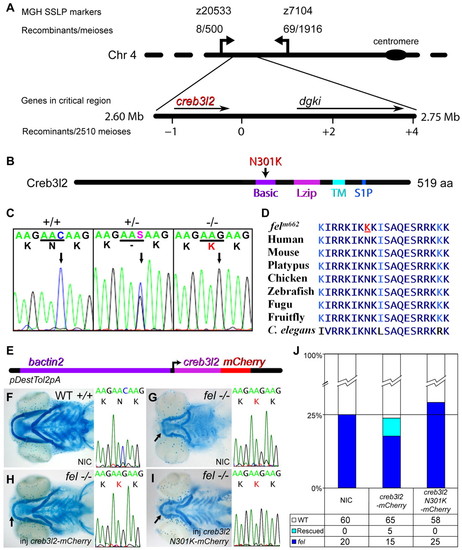
The feelgood line carries a missense mutation in creb3l2. (A) The feelgood mutation was mapped to chromosome 4 between SSLP markers z20533 and z7104. Novel SSLP markers with the indicated number of recombinants reduced the critical interval to a <50 kb region that contained two genes. The physical map of the critical region was based on the Zv7 assembly. (B) Schematic diagram of the Creb3l2 primary structure illustrating a missense N301K mutation in the DNA-binding basic motif. Basic, basic motif; Lzip, leucine zipper motif; TM, transmembrane domain; S1P, Site-1 protease recognition sequence. (C) Electropherograms of wild-type (+/+), heterozygous (+/-) and feelgood (-/-) genomic DNA. The arrow points to the C′G transversion that results in lysine for asparagine substitution at position 301 (N301K). (D) Comparison of the DNA-binding basic motif of creb3l2 across several species. The amino acid changed in feelgood mutants is underlined in red. (E) Schematic diagram of the Tol2kit-based construct containing mCherry-tagged creb3l2 under the ubiquitous β-actin (bactin2) promoter. (F–I) Alcian blue staining of cartilage elements at 5 dpf of non-injected (NIC) wild-type embryo (F); non-injected feelgood embryo (G); creb3l2-mCherry rescued feelgood mutant embryo (H); and mutant embryo injected with feelgood (N301K) creb3l2-Cherry that failed to rescue the phenotype (I). The chromatograms of sequences surrounding the feelgood lesion for the embryos shown in F–I have been included in the corresponding adjacent images. Arrows indicate an increased length of lower jaw in rescued mutant (H) compared with NIC feelgood control (G) or embryos injected with creb3l2 N301K (I). (J) Quantification of the number of mutant, wild-type and rescued phenotypes in the β-actin2 creb3l2-mCherry injection experiments. The feelgood genotype of these fish was confirmed by sequencing. |
|
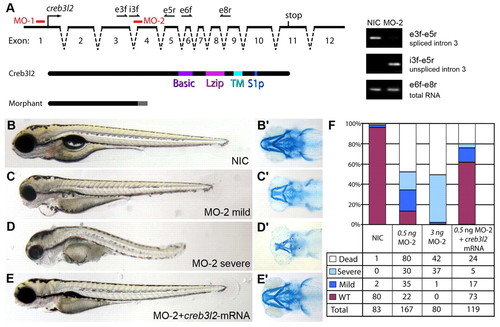
Decreased Creb3l2 function is responsible for the feelgood phenotype. (A) Schematic representation of the creb3l2 gene structure and positions of the two morpholinos MO-1 and MO-2 used in knockdown experiments. The names, position and orientation of specific primers used to evaluate the effectiveness of MO-2 are shown on top of the genomic locus diagram. Exonic primers are indicated by an ‘e’, intronic primers are indicated by an ‘i’. The predicted truncated form of the protein caused by MO-2 compared with the full-length wild-type protein is shown below (Morphant). PCR amplification products representing spliced and unspliced creb3l2 mRNA as compared with total creb3l2 mRNA in embryos injected with 3 ng MO-2 is shown on the right gel electrophoresis images. The primers used for amplification are shown on the right images. (B–E′) Live (lateral; B–E) and Alcian blue stained (ventral; B′–E′) images of 80-hpf embryos injected with 0.5 ng MO-2 (C,D), or MO-2 plus creb3l2-mRNA (E). NIC, non-injected control is shown in B. (F) Quantification of knockdown and mRNA rescue experiments indicates the percentage of embryos in the four phenotypic classes observed. PHENOTYPE:
|
|
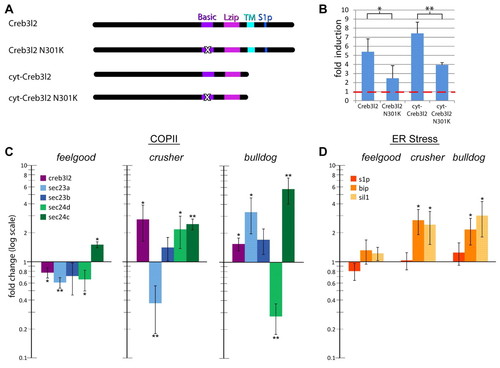
Expression of gene transcripts encoding COPII components is reduced in feelgood. (A) Schematic of zebrafish Creb3l2 expression constructs. ‘X’ in the basic domain marks the feelgood N301K mutation. (B) Luciferase reporter assays. Human fibroblasts were transfected with the indicated expression constructs and the firefly luciferase reporter containing 0.8 kb of the zebrafish sec23a promoter. Renilla luciferase control plasmid was also used as transfection efficiency control. Luciferase activities were compared with those of cells transfected with empty vector to determine fold induction. Red line indicates baseline activity adjusted to Renilla luciferase values. (C,D) qPCR analysis of the fold expression change of COPII-related transcripts creb3l2, sec23a, sec23b, sec24d and sec24c (C), and ER-stress-related transcripts s1p, bip and sil1 (D) in feelgood, crusher (sec23a) and bulldog (sec24d) mutants compared with wild types. RNA was extracted form whole embryos at 80 hpf. All results are normalized to β-actin and then to wild type. *P<0.05; **P<0.005. EXPRESSION / LABELING:
|
|
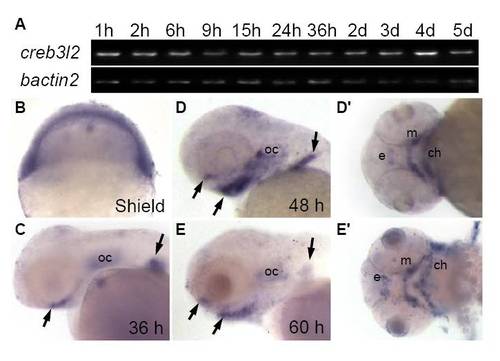
Expression of creb3l2 during zebrafish development. (A) RT-PCR analysis of creb3l2 expression with beta-actin as a loading control. (B-E′) Whole mount in situ hybridization of creb3l2 expression. Arrows indicate increased expression at the developing pharyngeal skeleton and fin between 36 and 60 hpf. C-E, lateral views, D′,E′ ventral views of D,E. Abbreviations: e, ethmoid plate; m, Meckel′s cartilage; ch, ceratohyal; oc, otic capsule. |