- Title
-
Hand2 ensures an appropriate environment for cardiac fusion by limiting Fibronectin function
- Authors
- Garavito-Aguilar, Z.V., Riley, H.E., and Yelon, D.
- Source
- Full text @ Development
|
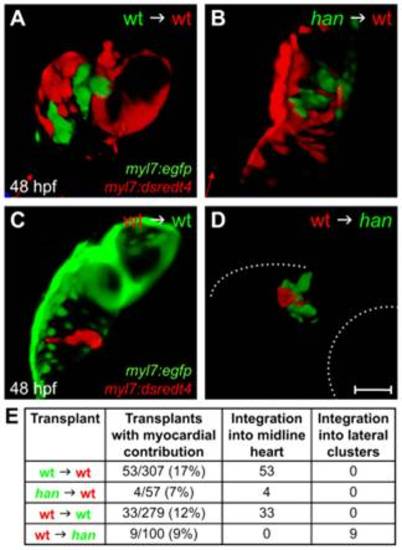
The role of hand2 in promoting cardiac fusion is not cell autonomous. (A-D) Confocal projections of mosaic hearts in live zebrafish embryos. (A-C) Lateral views, dorsal up. (D) Lateral view, dorsal down. (A,B) Wild-type host hearts expressing Tg(myl7:dsredt4) with integrated donor-derived cells expressing Tg(myl7:egfp) from wild-type (A) or han mutant (B) donors. Cells from wild-type or han mutant donors integrate indistinguishably into wild-type host hearts. (C,D) Wild-type (C) or han mutant (D) hosts expressing Tg(myl7:egfp) with integrated wild-type donor-derived cells expressing Tg(myl7:dsredt4). (D) In han mutant hosts, wild-type cells behave abnormally, remaining associated with lateral clusters of han mutant cardiomyocytes. Only the right-hand cluster is visible in this lateral view. Dotted lines indicate the embryo/yolk border (upper) and eye border (lower). Comparable clusters were observed in all nine chimeras examined, and no donor-derived cardiomyocytes were found outside of the clusters or at the midline. Scale bar: 100 μm. (E) Summary of results, indicating the ratio of hosts with donor-derived cardiomyocytes to all host embryos screened and the integration of donor-derived cardiomyocytes into the midline heart or lateral clusters. |
|
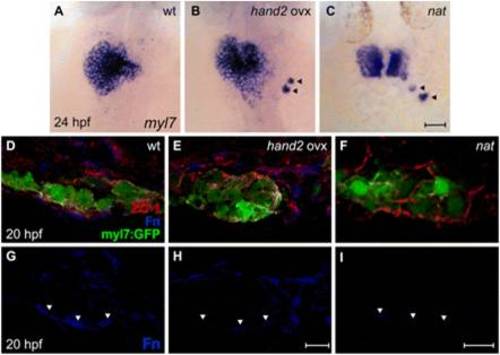
hand2 overexpression reduces Fn deposition. (A-C) In situ hybridization for myl7. Dorsal views, anterior up. Zebrafish embryos overexpressing hand2 (B) and nat mutant embryos (C) exhibit scattered cardiomyocytes (arrowheads), as well as delayed cardiac fusion. (D-I) Transverse confocal sections of the left lateral mesoderm in embryos expressing Tg(myl7:egfp) (green). Dorsal is up. Immunofluorescence detects ZO-1 (red) and Fn (blue). In contrast to the wild-type monolayer (D), cardiomyocytes are disorganized and multilayered in hand2-overexpressing embryos (E) and nat mutants (F). Furthermore, the deposition of Fn basal to the myocardium (G, arrowheads) is significantly reduced in hand2-overexpressing embryos (H; also see Fig. S2 in the supplementary material) and absent in nat mutants (I). Scale bars: 50 μm in A-C; 10 μm in D-I. |
|
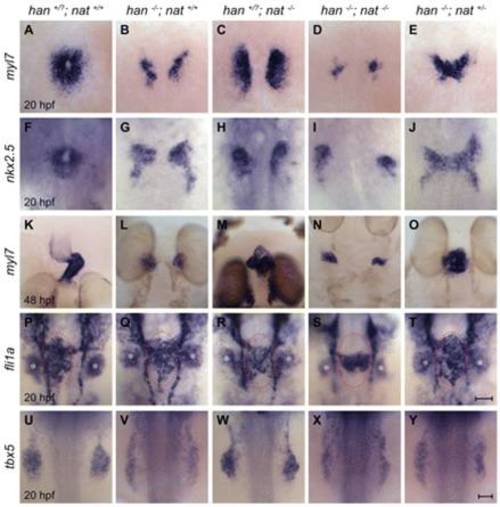
Reduction of fn1 function rescues cardiac fusion in hand2 mutants. In situ hybridizations for the indicated genes. Dorsal views, anterior up. Zebrafish embryos were genotyped by PCR following imaging, and all phenotypes were found to be consistent; for example, the rescue observed in E was seen in a total of 18 embryos from three independent clutches. (A-J) Cardiac fusion progresses in wild-type and han-/-nat+/- embryos, whereas cardia bifida is present in han, nat and han-/-nat-/- embryos. (K-O) In contrast to the wild-type midline heart, a cardiac cone forms in han-/-nat+/- embryos, a bifurcated heart forms in nat mutants, and cardia bifida is seen in han and han-/-nat-/- embryos. The nat cardiac phenotype varies, ranging from a midline heart to cardia bifida (Trinh and Stainier, 2004). (P-T) Endocardial sheet formation in wild-type and han-/-nat+/- embryos contrasts with the dysmorphic endocardium in han, nat and han-/-nat-/- embryos. Red ovals mark the extent of anterior-posterior spreading of the endocardial sheet in wild-type and han-/-nat+/- embryos; this spreading appears inadequate in han, nat and han-/-nat-/- embryos, either due to morphogenesis or cell number defects. In addition to its endothelial expression, fli1a is expressed in branchial arch mesenchyme (asterisks). (U-Y) The failure of pectoral fin mesenchyme development in han embryos is not modified by altering nat function, in contrast to the normal fin mesenchyme observed in wild-type or nat embryos. Scale bars: 50 μm. |
|
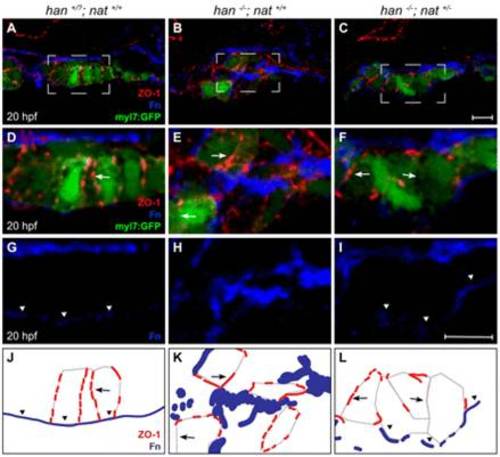
Tissue architecture and Fn localization improve without rescue of polarity. (A-I) Transverse confocal sections of the right lateral mesoderm in zebrafish embryos expressing Tg(myl7:egfp) (green). Dorsal is up. ZO-1 (red) and Fn (blue) are detected by immunofluorescence. The boxed regions in A-C are shown at higher magnification in D-I. Scale bars: 10 μm. (J-L) Schematics of the cardiomyocyte-associated localization of ZO-1 (red) and Fn (blue) as shown in D-I. Wild-type cardiomyocytes are organized in a cohesive layer (A) and exhibit lateral localization of ZO-1 (D,J, arrows) and basal deposition of Fn (D,G,J, arrowheads). In han embryos, tissue architecture is aberrant (B) and myocardial apicobasal polarity is not evident (E,H,K). ZO-1 is diffuse or absent (E,K, arrows) and Fn deposition is disorganized (E,H,K). In han-/-;nat+/- embryos, myocardial cohesion (C) and basal Fn localization (F,I,L, arrowheads) are improved, but polarity is not consistently rescued: localization of ZO-1 is either lateral or absent (F,L, arrows). |
|
Heightened levels of fn1 expression in han mutants. (A,B) In situ hybridization for myl7 illustrates the han mutant phenotype at the stage when embryos were collected for RNA extraction and microarray analysis. Dorsal views, anterior up. Scale bar: 50 μm. (A) Wild-type cardiomyocytes have initiated cardiac fusion with contacts between posterior subsets of contralateral cells and are proceeding to complete formation of the cardiac cone with the merger of anterior subsets of contralateral cells. (B) By contrast, han mutant embryos exhibit two separated populations of cardiomyocytes. (C,D) Bar charts indicating levels of gene expression, relative gapdh, as detected by qRT-PCR in wild-type and han mutant embryos. Values represent the mean (±s.e.m.) from triplicate samples. (C) Microarray analysis revealed a 2.0-fold upregulation of fn1 in han mutants (see Table S1), and validation of this result by qRT-PCR indicated that fn1 expression is increased 1.6-fold in han mutants. (D) Expression of hand2 was undetectable by qRT-PCR in han mutants, whereas microarray analysis indicated a 5.3-fold downregulation of hand2 in han mutants (see Table S1). PCR primers used were: fn1, 52-GAATCCCCGAGCCAATCAA-32 and 52-TTTTATGTTTCCCCGTTCCTT-32 hand2, 52-TGCGCTCGAATCAACAAATG-32 and 52-TATACCCTACGGCGAACCACA-32 gapdh, 52-CCCCACCCCCAATGTCTCT-32 and 52-AACCTGGTGCTCCGTGTATCC-32. |
|
Quantification confirms reduced Fn deposition basal to the myocardium in embryos overexpressing hand2. (A) Representative transverse confocal section in an embryo expressing Tg(myl7:egfp) (green) exemplifies the method for quantification of Fn immunofluorescence (blue). Scale bar: 10 μm. For each image, we used ImageJ software to select a cardiac region of interest (r.o.i.) (solid rectangle) basal to a group of at least three cardiomyocytes and then measured the mean pixel intensity of the r.o.i. in the blue channel. Similarly, we measured the mean pixel intensity in a comparably sized r.o.i. (dashed rectangle) along the exterior cortex of the embryo. Measurements were taken from three consecutive confocal images per cryosection in three different cryosections per embryo, and at least three embryos were sampled for each condition. (B) Bar chart comparing the levels of Fn deposition in terms of mean pixel intensity (±s.e.m.) for each r.o.i. measured. Asterisk indicates statistically significant difference from wild type (P<0.0001, two-tailed unpaired Student′s t-test). In embryos overexpressing hand2, deposition of Fn adjacent to the cardiomyocytes is reduced <85%, whereas cortical deposition of Fn is relatively unaffected. (C) Bar chart comparing levels of Fn deposition adjacent to the cardiomyocytes, normalized relative to cortical Fn levels. Values represent the mean ratio (±s.e.m.) for each condition; asterisk indicates statistically significant difference from wild type (P<0.0001, two-tailed unpaired Student′s t-test). (D) Bar chart indicating levels of fn1 expression, relative to hprt expression, as detected by qRT-PCR in wild-type embryos and in embryos overexpressing hand2. Values represent the mean (±s.e.m.) from triplicate samples. fn1 expression is decreased 4-fold in embryos overexpressing hand2. PCR primers used were: fn1, 52-GAATCCCCGAGCCAATCAA-32 and 52-TTTTATGTTTCCCCGTTCCTT-32 hprt, 52-ATCCGCCTCAAGAGTTACCAA-32 and 52-TCTAGCAGCGTTTTCATCGTT-32. |
|
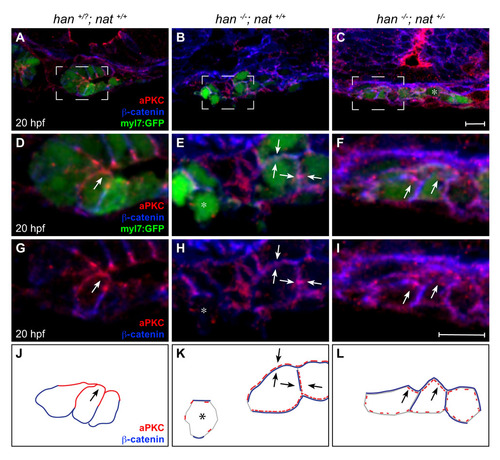
Poor rescue of myocardial apicobasal polarity in han-/-;nat+/- embryos. (A-I) Transverse confocal sections of the right lateral mesoderm in embryos expressing Tg(myl7:egfp) (green). Dorsal is up. Immunofluorescence detects aPKC (red) and β-catenin (blue). Boxed regions in A-C are shown at higher magnification in D-I. At this stage, the wild-type myocardium has begun the asymmetric involution that displaces a medial portion of the myocardial epithelium dorsally (Rohr et al., 2008). Scale bars: 10 μm. (J-L) Cartoons schematize cardiomyocyte-associated localization of aPKC (red) and β-catenin (blue) shown in D-I. Wild-type cardiomyocytes are organized in a cohesive layer (A) and exhibit apical enrichment of aPKC (D,G,J, arrow) and basolateral enrichment of β-catenin (D,G,J). In han embryos, the small clusters of cardiomyocytes (B) exhibit irregular localization of aPKC and β-catenin, either diffusely around the cell (E,H,K, arrows) or nearly absent (E,H,K, asterisk). Only slight improvements are seen in han-/-;nat+/- embryos: aPKC is either distributed diffusely (C, asterisk) or partially enriched apically (F,I,L, arrows), and β-catenin is either distributed diffusely or partially enriched laterally (F,I,L). We note that previous work indicated an absence of aPKC localization in han mutant cardiomyocytes (Trinh et al., 2005). We suspect that the diffuse and irregular aPKC localization observed here (E,H,K) reflects increased sensitivity of our current protocol, which uses a different fixation method and thinner sections. |
|
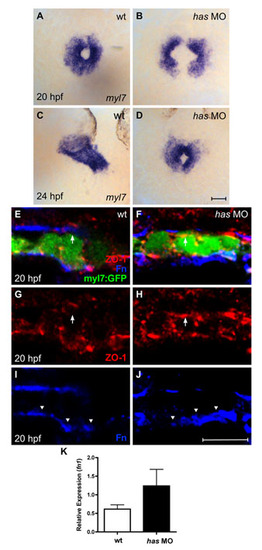
Loss of apkci function does not disrupt basal deposition of Fn. (A-D) In situ hybridization for myl7 comparing embryos injected with an anti-apkci morpholino (has MO) with their wild-type siblings. Dorsal views, anterior up. Scale bar: 50 μm. Although cardiac fusion proceeds in embryos lacking apkci function, heart tube extension is arrested (Horne-Badovinac et al., 2001; Peterson et al., 2001; Rohr et al., 2006). (E-J) Transverse confocal sections of the left lateral mesoderm in embryos expressing Tg(myl7:egfp) (green). Dorsal is up. Immunofluorescence detects ZO-1 (red) and Fn (blue). Scale bar: 10 μm. Wild-type cardiomyocytes exhibit lateral localization of ZO-1 (E,G) and basal deposition of Fn (E,I, arrowheads). By contrast, has morphant cardiomyocytes exhibit defective apicobasal polarity (Rohr et al., 2006): for example, ZO-1 is diffusely localized (F,H), including localization in the apical domain, where it is not found in wild-type cardiomyocytes (E-H, arrows). Despite the disruption of some aspects of apicobasal polarity, Fn deposition remains restricted to the basal domain of has morphant cardiomyocytes (F,J, arrowheads). Altogether, the has morphant phenotype shares many similarities with the han-/-;nat+/- phenotype. In each case, cardiac fusion proceeds, but heart tube extension arrests; additionally, the myocardium is a relatively aligned layer that exhibits basal Fn deposition despite its other apicobasal polarity defects. (K) Bar chart indicates levels of fn1 expression, relative to hprt expression, as detected by qRT-PCR in wild-type and has morphant embryos. Values represent the mean (±s.e.m.) from triplicate samples. fn1 expression is increased 2-fold in has morphants, comparable to the increase observed in han mutants (see Fig. S1C). However, our qRT-PCR data monitor fn1 expression in whole embryos, rather than in the specific locations relevant to cardiac fusion. In this regard, it is notable that the global increase in fn1 expression in has morphants does not block cardiac fusion (D) and that these embryos also exhibit normal basal deposition of Fn (J). PCR primers used were: fn1, 52-GAATCCCCGAGCCAATCAA-32 and 52-TTTTATGTTTCCCCGTTCCTT-32 hprt, 52-ATCCGCCTCAAGAGTTACCAA-32 and 52-TCTAGCAGCGTTTTCATCGTT-32. |