- Title
-
Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos
- Authors
- Lindeman, L.C., Winata, C.L., Aanes, H., Mathavan, S., Alestrom, P., and Collas, P.
- Source
- Full text @ Int. J. Dev. Biol.
|
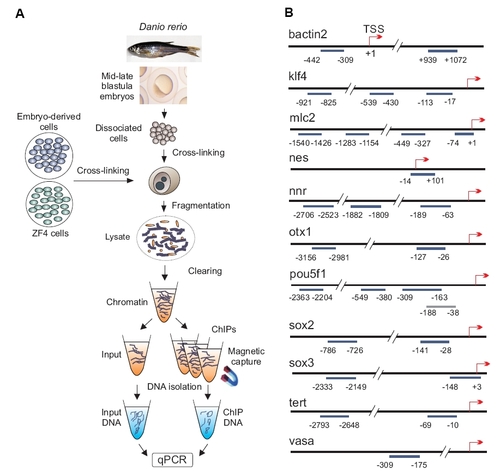
Chromatin immunoprecipitation (ChIP) assay and map of genomic regions examined in this study. (A) Post-translational histone modifications in MBT+ stage embryos, ZF4 cells and MBT+ embryo-derived cultured cells were examined by ChIP-qPCR. (B) Genomic regions examined by ChIP on indicated genes. Numbers indicate the position of amplicons relative to the TSS (+1). |
|
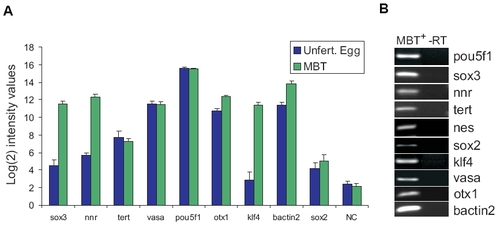
Expression pattern of selected genes in zebrafish embryos. (A) Data extracted from Agilent microarray analyses of gene expression in unfertilized eggs and MBT embryos. Data was generated from 3 biological replicates for both stages, with 3 and 4 arrays for unfertilized egg and MBT stages, respectively. Signals were quantile-normalized, multiple probes for each gene were aggregated using the median, and mean±SD calculated based on log2-transformed values. Negative controls are the mean of the replicates of the median of 150 different negative control probes scattered throughout the array. (B) RT-PCR analysis of expression of indicated genes in MBT+ embryos; -RT, PCR without reverse transcription. |
|
Embryonic genes are repressed and trimethylated on H3K9 and H3K27 in somatic tissue. (A) RT-PCR analysis of expression of indicated genes in muscle biopsies from three different fish; -RT, PCR without reverse transcription. (B,C) ChIP analysis of indicated histone modifications on the promoters of indicated genes. In (C), insets highlight H3K9ac and H4ac occupancy on mlc2 and bactin2. |
|
Histone modification and gene expression patterns in cultured embryo- derived cells. (A) Cells isolated from MBT+ embryos were cultured for 24 h and histone PTMs examined as in Figure 3. (B) RT-PCR analysis of gene expression in embryo-derived cells cultured for 1, 2 or 5 days. -RT, PCR without reverse transcription on RNA isolated from Day 1 cultured cells. |
|
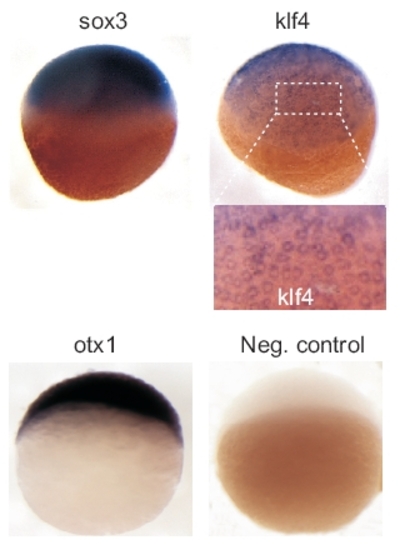
Whole-mount in situ hybridization analysis of expression of indicated genes in MBT+ stage embryos. Enlargement of klf4 hybridization pattern (boxed area) shows mosaic expression between blastomeres. A negative control otx1b hybridization using a sense otx1b probe is also shown. |
|
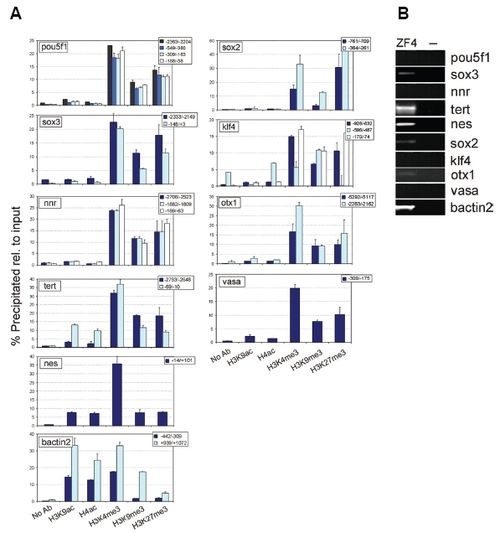
Histone modification patterns on promoters of developmentally-regulated genes in the ZF4 cell line. (A) ChIP-qPCR analysis of histone PTMs on indicated genomic sites (relative to the TSS). (B) RT-PCR analysis of expression of indicated genes; “-“ refers to a control PCR without reverse transcription. |