- Title
-
In vivo correction of a Menkes disease model using antisense oligonucleotides
- Authors
- Madsen, E.C., Morcos, P.A., Mendelsohn, B.A., and Gitlin, J.D.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
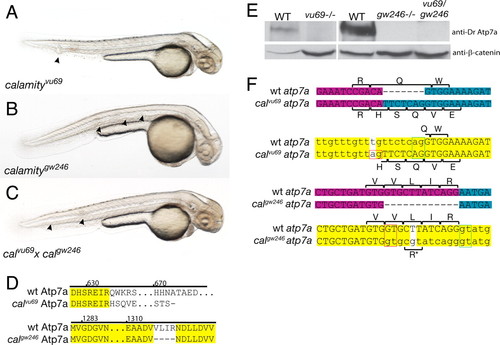
Comparison of phenotype, genotype, and protein expression for two alleles of calamity. (A) calamityvu69 , characterized in ref. 7, displays the copper-deficient phenotype of loss of pigmentation, notochord abnormalities (arrowheads), and an enlarged ventricle and is due to a loss of function of atp7a. (B) calamitygw246 , a second allele of atp7a, has the same phenotype. (C) calgw246 fails to complement calvu69 , confirming allelism. (D) Amino acid sequence changes caused by the mutations. calvu69 results in a frameshift and truncation of the C terminus of the protein. calgw246 contains an in-frame deletion in the highly conserved ATPase domain of Atp7a. (E) Immunoblot analysis using an antibody made to a C-terminal peptide from zebrafish Atp7a shows complete loss of WT protein in calvu69 , calgw246 , and in the compound heterozygote. (F) The mRNA and genomic sequences of calvu69 (Upper) and calgw246 (Lower), highlighting the mutation and subsequent changes in amino acid sequence due to missplicing of exons through the aberrant use of mutant splice sites (red boxes). Exon boundaries in the mRNA are illustrated by a change in color from magenta to blue. In the genomic sequence, introns are in lowercase and the mutations are the bases with the white background. The green boxes delineate the WT splice donor/acceptor pair and red boxes the mutant pair. For calgw246 , a small percentage of transcripts splice at the WT splice junction retaining the nonsynonymous mutation, a Leu to Arg amino acid substitution (*). |
|
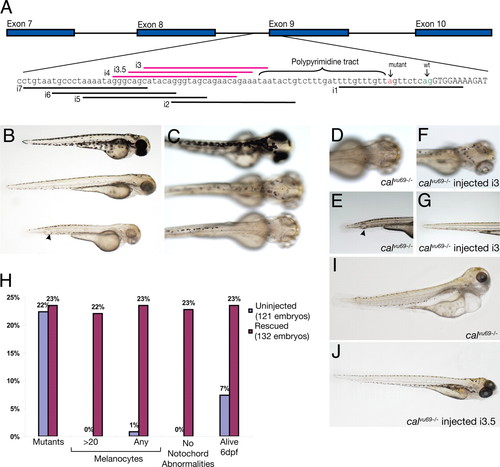
Morpholinos rescue the phenotype of calvu69 embryos. (A) Schematic and sequence information for the design of morpholinos directed toward the rescue of the mutant splicing in calvu69 . The WT exon is uppercase. The mutation is highlighted in red, and the WT splice acceptor is in green. Each morpholino-binding site is indicated by a line located 52 to the polypyrimidine tract. Morpholinos that cause phenotypic rescue are highlighted in pink. Morpholino i1 served as a control to demonstrate that blocking of proper splicing at this exon–exon boundary would result in the calamity phenotype (data not shown). (B and C) Side-by-side comparisons of zebrafish 48 h after fertilization. (Top) WT sibling from calvu69 intercross clutch. (Bottom) calvu69/vu69 -mutant embryo. (Middle) calvu69/vu69 embryo rescued by injection of morpholino i3. (D and E) Close-up view of calvu69/vu69 -uninjected embryo showing lack of melanin pigmentation over head region and notochord abnormalities (arrowhead). (F and G) Close-up view of morpholino i3-rescued calvu69/vu69 embryo showing restoration of melanin pigmentation and a notochord absent of defects. (H) Quantification of the extent of rescue. A single group of embryos derived from several clutches, half injected with rescuing morpholino (red bars; uninjected, blue bars), was scored for the presence of any or a significant number of melanocytes (>20) and the presence of notochord defects. The scale of the graph reflects the expected 25% ratio of homozygous mutants derived from the heterozygous intercross. (I and J) Six days after fertilization, embryos were either uninjected (I) or injected (J) with rescuing morpholino i3.5. Rescued embryos maintain near-normal body morphology, whereas unrescued embryos swell, possibly because of decreased extracellular matrix integrity due to the loss of lysyl oxidase activity. |
|
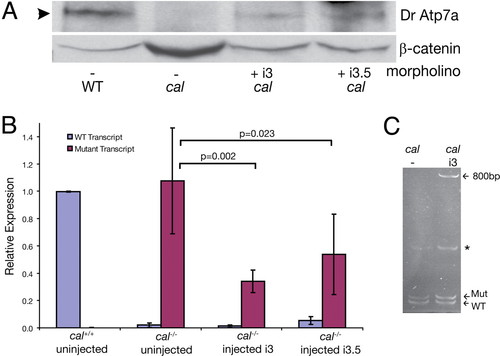
Rescue morpholinos cause restoration of full-length protein without detectable changes in mRNA. (A) Phenotypic rescue by morpholinos i3 and i3.5 is accompanied by the restoration of the WT protein. Forty-eight hours after fertilization, lysates from embryos of the indicated genotype were run on a SDS/6% PAGE gel and blotted for zebrafish Atp7a by using an antibody generated to the zebrafish protein. β-catenin was used as a loading control. Morpholino injection restored protein to 30–45% of WT levels in this experiment. (B) qRT-PCR using primers specific for WT and mutant atp7a transcript were used to measure the amounts of each in single embryos. Normalization of WT transcript was to cal +/+ embryos and mutant transcript to cal-/- embryos. Error bars indicate one standard deviation around the mean. (C) Polyacrylamide gel of RT-PCR products reveals a band containing 800 bp of intronic sequence caused by morpholino injection. This band was sequenced and represents use of a cryptic 32 splice site located within the intron. Also seen are mutant and WT transcripts in uninjected and rescued embryos. A nonspecific band was present in both uninjected and injected mutant embryos (*). EXPRESSION / LABELING:
|