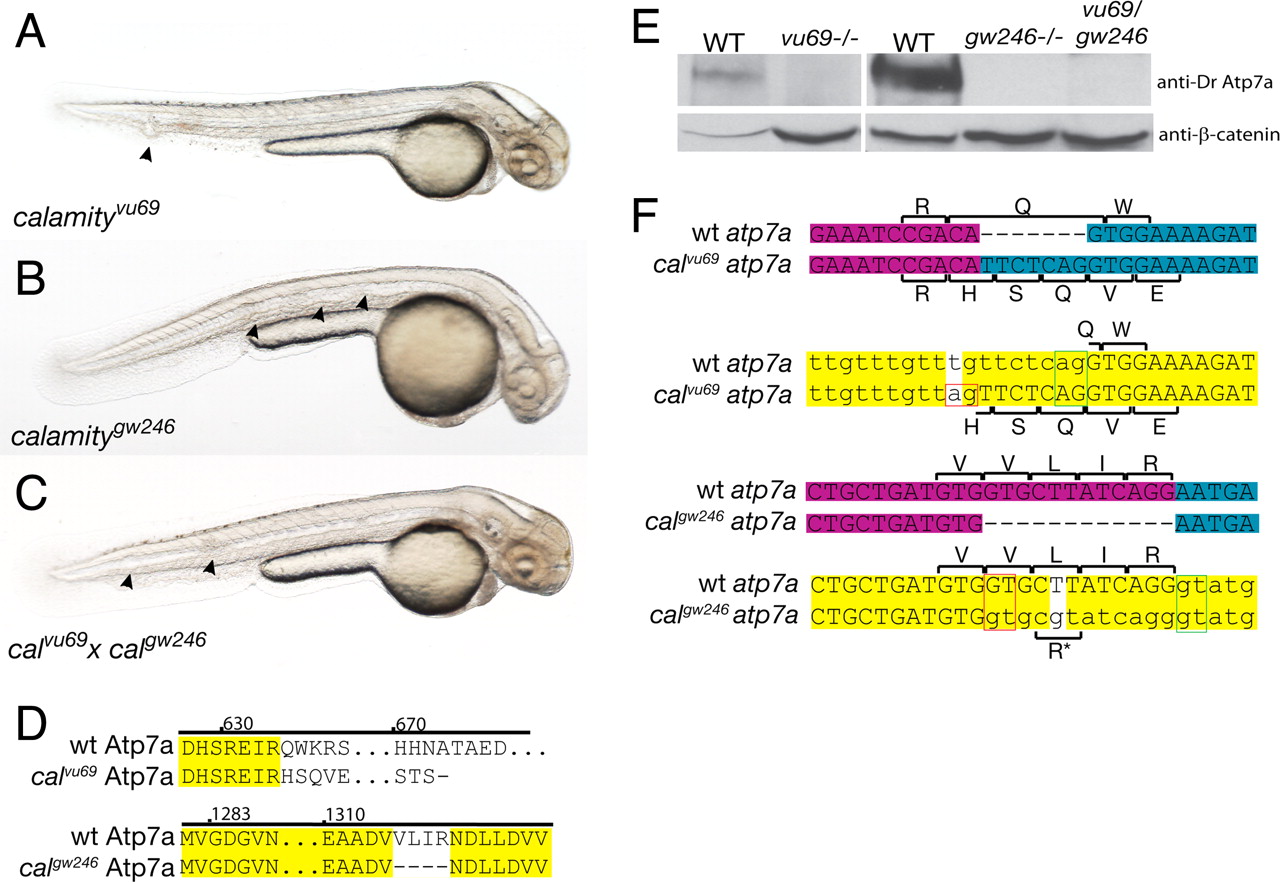
Fig. 1
Comparison of phenotype, genotype, and protein expression for two alleles of calamity. (A) calamityvu69 , characterized in ref. 7, displays the copper-deficient phenotype of loss of pigmentation, notochord abnormalities (arrowheads), and an enlarged ventricle and is due to a loss of function of atp7a. (B) calamitygw246 , a second allele of atp7a, has the same phenotype. (C) calgw246 fails to complement calvu69 , confirming allelism. (D) Amino acid sequence changes caused by the mutations. calvu69 results in a frameshift and truncation of the C terminus of the protein. calgw246 contains an in-frame deletion in the highly conserved ATPase domain of Atp7a. (E) Immunoblot analysis using an antibody made to a C-terminal peptide from zebrafish Atp7a shows complete loss of WT protein in calvu69 , calgw246 , and in the compound heterozygote. (F) The mRNA and genomic sequences of calvu69 (Upper) and calgw246 (Lower), highlighting the mutation and subsequent changes in amino acid sequence due to missplicing of exons through the aberrant use of mutant splice sites (red boxes). Exon boundaries in the mRNA are illustrated by a change in color from magenta to blue. In the genomic sequence, introns are in lowercase and the mutations are the bases with the white background. The green boxes delineate the WT splice donor/acceptor pair and red boxes the mutant pair. For calgw246 , a small percentage of transcripts splice at the WT splice junction retaining the nonsynonymous mutation, a Leu to Arg amino acid substitution (*).