- Title
-
Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways
- Authors
- Yamaguchi, M., Tonou-Fujimori, N., Komori, A., Maeda, R., Nojima, Y., Li, H., Okamoto, H., and Masai, I.
- Source
- Full text @ Development
|
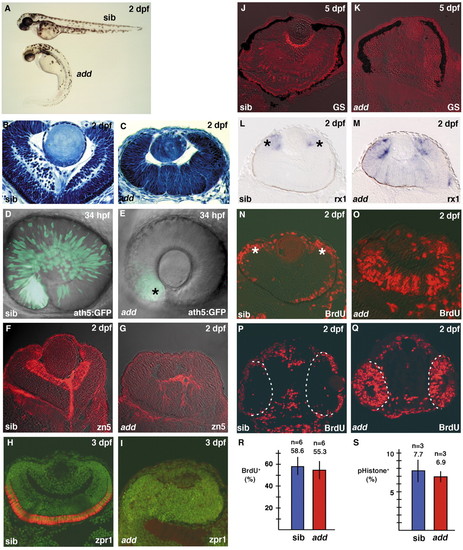
Retinal mitotic cells fail to exit from the cell cycle in add mutant embryos. (A) Morphology of add mutant and wild-type sibling embryos. (B,C) Plastic sections of wild-type (B) and add mutant (C) retinas. add mutant embryos show a markedly multifolded retinal epithelium. (D,E) ath5:GFP expression in wild-type (D) and add mutant (E) retinas. ath5:GFP expression is markedly delayed in add mutant embryos and very faint in the ventronasal retina (E, asterisk). (F,G) Labeling of wild-type (F) and add mutant (G) retinas with the zn5 antibody, which stains retinal ganglion cells (red). (H,I) Labeling of wild-type (H) and add mutant (I) retinas with the zpr1 antibody, which stains double cone photoreceptors (red). All nuclei were counterlabeled with Sytox Green (green). (J,K) Labeling of wild-type (J) and add mutant (K) retinas with the anti-glutamine synthetase antibody, which stains Müller glial cells (red). (L,M) In situ hybridization of wild-type (L) and add mutant (M) retinas with rx1 RNA probe. rx1 expression is observed in nearly the entire neural retina of add mutants, whereas it is downregulated and localized within a proliferating region called the CMZ (L, asterisks) in wild type. (N,O) BrdU labeling of wild-type (N) and add mutant (O) retinas. BrdU-positive cells are located within the CMZ (N, asterisks) and a part of the outer photoreceptor layer in wild type, whereas many retinal cells incorporate BrdU even in the central retina of add mutant embryo. (P,Q) BrdU labeling of wild-type (P) and add mutant (Q) heads. Broken white lines show the interface between the brain and retina at 24 hpf. (R) The percentage of the number of BrdU-positive cells to total number of cells in wild-type (blue bar) and add mutant (red bar) retinas. (S) The percentage of the number of mitotic cells labeled with anti-phosphorylated Histone H3 antibody in wild-type (blue bar) and add mutant (red bar) retinas at 27 hpf. EXPRESSION / LABELING:
|
|
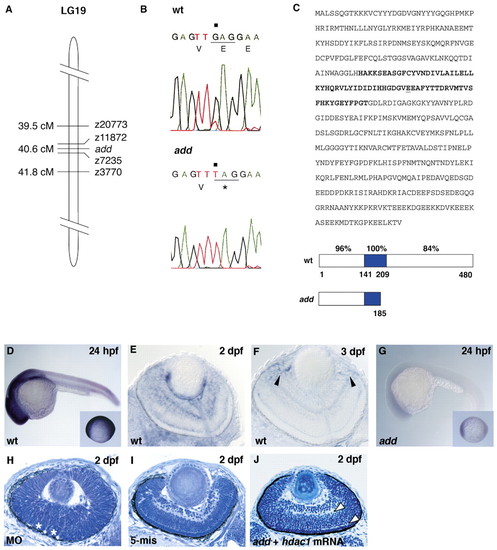
add gene encodes Hdac1. (A) Schematic drawing of LG19. (B) Sequencing of hdac1 DNA isolated from wild type and add mutants. G to T transversion (black squares) replaces 185 E with stop codon in add mutants. (C) Amino acid sequence of Hdac1 and its predicted structure of wild type and add mutant. Nonsense mutation occurs at 185 E (underline) within the Hdac catalytic region (bold letters in sequence, blue in schematic drawing below). Percentage of identical amino acids in zebrafish and human Hdac1 is indicated in each domain. (D-G) Expression of hdac1 mRNA in wild-type (D-F) and add mutant (G) embryos. Arrowheads (F) indicate the expression in the CMZ. Insets (D and G) show embryos at 10 hpf. (H) Retina of embryos injected with hdac1 morpholino oligos. Asterisks indicate folded retinal epithelium. (I) Retina of embryos injected with five mismatch control morpholino oligos. Retinal lamination is normal. (J) add mutant retinas expressing hdac1 mRNA. The formation of the inner and outer plexiform layers are evident (arrowheads). EXPRESSION / LABELING:
|
|
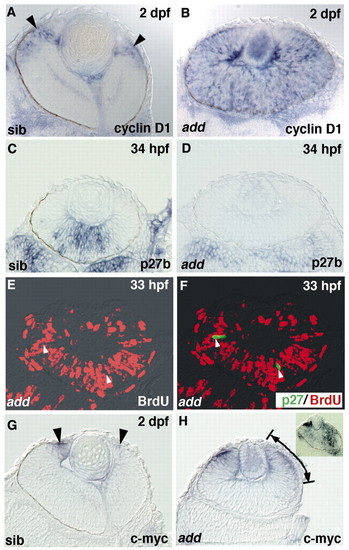
Hdac1 functions upstream of the interaction between cyclin D1 and p27. (A,B) cyclin D1 expression in wild-type (A) and add mutant (B) retinas. Almost all retinal cells express cyclin D1 in add mutants, while in wild-type retinas, cyclin D1 expression level decreases except in the CMZ (arrowheads). (C,D) p27b expression in wild-type (C) and add mutant (D) retinas. p27b expression is absent in the add mutant retinas. (E,F) add mutant retinas expressing Myc-tagged Xenopus p27 labeled with anti-Myc antibody (F, green) and anti-BrdU antibody (red). The introduction of Xenopus p27 inhibits BrdU incorporation in the add mutant retinas (arrowheads). (G,H) cmyc expression in wild-type (G) and add mutant retinas (H). cmyc is expressed in the peripheral CMZ (arrowheads) in wild-type retinas, while cmyc expression spreads centrally in add mutant retinas (arrows/lines). cmyc expression in wild-type retina treated with 1200 nM TSA (H, inset) spreads over a large region. |
|
Notch/Hes signaling pathway is activated in the add mutant retina. (A) Lateral view of 34 hpf wild-type retinas expressing ΔN-Tcf3. A confocal image of a 4.5 µm plane is shown. Although ath5:GFP expression (green) progresses to the large region of the neural retina in this stage (see Fig. 1D), ath5:GFP spreads to the dorsonasal retina in this plane. Some of ΔN-Tcf3-expressing cells (red) are ath5:GFP positive (arrows, yellow). (B) A confocal image of lateral view of 34 hpf add mutant retinas expressing ΔN-Tcf3. Very faint ath5:GFP expression is detected in the ventronasal retina (green, arrowhead). Cells expressing ΔN-Tcf3 (red) in the dorsal retina do not express ath5:GFP (arrows). (C-D) In situ hybridization of wild-type (C) and add mutant (D) retinas with her4 RNA probe. In the wild-type retina, her4 expression is localized in the central part of the CMZ (lines and arrows), which corresponds to the intermediate zone between retinal stem cells (asterisks) and differentiated neurons. her4 expression is also observed in a part of the outer layer, where neurogenesis occurs at this stage. By contrast, her4 expression remains in the large region of the neural retina in the add mutant (D). (E,F) In situ hybridization of wild-type (E) and wild-type retinas expressing NICD (F) with her4 RNA probe. her4 expression is upregulated in NICD-expressing retinas. (G,H) In situ hybridization of mib (G) and add; mib double mutant (H) retinas with her4 RNA probe. her4 expression is not observed in either case. (I,J) In situ hybridization of wild-type (I) and wild-type retinas expressing Δ47-ß-catenin (J) with her4 RNA probe. her4 is expressed in the CMZ and the presumptive outer layer, the latter of which is undulated because of Δ47-ß-catenin-induced folding of the neural retina. EXPRESSION / LABELING:
|
|
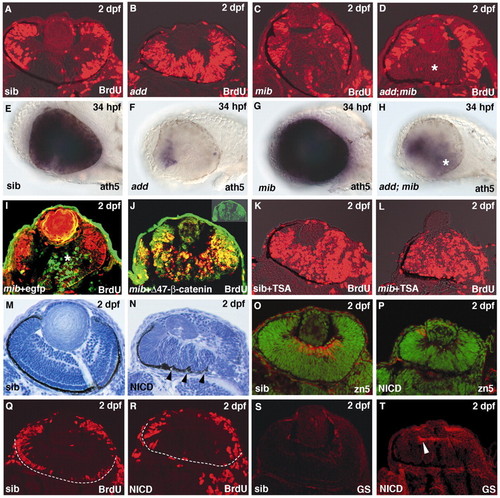
Notch signaling inhibits both cell-cycle exit and neurogenesis in the retina. (A-D) BrdU labeling in wild-type (A), add (B), mib (C) and add; mib mutant retinas. BrdU incorporation is suppressed in the central region of the add; mib double mutant retina (D, asterisk). (E-H) ath5 expression in wild-type (E), add (F), mib (G) and add; mib (H) mutant retinas. ath5 expression is severely inhibited in add mutant retinas, is enhanced in mib mutant retinas and is partially restored in the add; mib double mutant retinas (H, asterisk). (I,J) BrdU labeling of mib mutant injected with the DNA construct pCS2[hsp:EGFP] (I) and with pCS2[hsp:Δ47-ß-catenin-GFP] (J). Cells expressing Δ47-ß-catenin (green, inset in J) are BrdU-positive (red), even in the central region of mib mutant retinas (J), while cells expressing EGFP are BrdU negative in the central region of mib mutant retinas (I). (K,L) BrdU labeling of wild-type retina treated with TSA (K) and mib mutant retina treated with TSA (L). Treatment with 400 nM TSA from 10 hpf induces more severe phenotypes than that from 14 hpf shown in Fig. 5F. (M,N) Plastic sections of retinas of wild-type retina (M) and wild-type retina expressing NICD (N). Wild-type retina expressing NICD is folded in the same way as the add mutant retina (arrowheads). (O,P) Labeling of wild-type (O) and NICD-expressing (P) retinas with zn5 antibody, which labels retinal ganglion cells (red). Nuclei were counterlabeled with Sytox Green (green). (Q,R) BrdU labeling of wild-type (Q) and NICD-expressing retinas (R). In both cases, BrdU incorporation is not observed in the central retina. Broken white lines show the interface between the brain and retina. (S,T) Labeling of wild-type (S) and NICD-expressing retinas (T) with anti-glutamine synthetase antibody, which labels Müller glia (red). Glial cells differentiate precociously in the NICD-expressing retina (arrowhead) EXPRESSION / LABELING:
|

Unillustrated author statements EXPRESSION / LABELING:
|